Original research |
Peer reviewed |
Prairie dog (Cynomys ludovicianus) is not a host for porcine reproductive and respiratory syndrome virus
El perrito de la pradera (Cynomys ludovicianus) no es un huésped para el virus del síndrome reproductivo y respiratorio porcino
Le chien de prairie (Cynomys ludovicianus) n’est pas un hôte pour le virus du syndrome reproducteur et respiratoire porcin
Rodney B. Baker, DVM, MS; Wanqin Yu; Martha Fuentes, DVM, PhD; Craig R. Johnson, PhD; LaRae Peterson; Kurt Rossow, DVM, PhD; C. Scanlon Daniels, DVM; Angela M. Daniels, DVM; Dale Polson, DVM, PhD; Michael P. Murtaugh, PhD
RBB: Department of Population Health and Pathobiology, North Carolina State University, Raleigh, North Carolina. WY, MF, CRJ, LRP, MPM: Department of Veterinary and Biomedical Sciences, University of Minnesota, St Paul, Minnesota. KR: Department of Veterinary Population Medicine, University of Minnesota, St Paul, Minnesota. CSD, AMD: Circle H Animal Health, Dalhart, Texas. DP: Boehringer Ingelheim Vetmedica, Ames, Iowa. Corresponding author: Dr Michael Murtaugh, Department of Veterinary and Biomedical Sciences, University of Minnesota, 1971 Commonwealth Avenue, St Paul, MN 55108; Tel: 612-625-6735; Fax: 612-625-5203; E-mail: murta001@umn.edu.
Cite as: Baker RB, Yu W, Fuentes M, et al. Prairie dog (Cynomys ludovicianus) is not a host for porcine reproductive and respiratory syndrome virus. J Swine Health Prod. 2007;15(1):22–29.
Also available as a PDF.
SummaryObjective: To determine if the prairie dog is a biological host for porcine reproductive and respiratory syndrome virus (PRRSV). Materials and methods: Sixteen wild-caught prairie dogs were inoculated intraperitoneally with a high-titered mixture of wild-type local PRRSV strains, and seven animals were uninoculated controls. Serum and tissues were collected at 3- to 7-day intervals through 28 days for reverse transcriptase-polymerase chain reaction (RT-PCR) analysis of viral RNA and determination of an anti-PRRSV immune response, using a novel ELISA to measure specific prairie dog IgG responses to nine viral polypeptides. Results: A variety of tissues were negative for PRRSV RNA at all time points in both treated and control animals. Clinical signs were unremarkable and no histopathological lesions of PRRS were observed. Seroconversion was not observed in any animal over the 28-day study time course. Individual variation in background antibody levels and RT-PCR results were observed. Implications: Prairie dogs do not support replication of PRRSV and are not a reservoir of the virus. Quantitative RT-PCR values for PRRSV in serum and tissue samples overlap with negative background values near the limits of detection, increasing the risk of false-positive interpretations. Methods, materials, and resources for diagnostic investigation of the prairie dog are now available to the veterinary community. | ResumenObjetivo: Determinar si el perrito de la pradera es un huésped biológico para el virus del síndrome reproductivo y respiratorio porcino (PRRSV por sus siglas en inglés). Materiales y métodos: Se inocularon intraperitonealmente dieciséis perritos de la pradera salvajes con una mezcla con altos títulos de cepas de campo locales de PRRSV, y siete animales fueron los controles no inoculados. Se recolectaron sueros y tejidos en intervalos de 3- a 7- días por 28 días para el análisis de la reacción en cadena de polimerasa por transcripta reversa (RT-PCR por sus siglas en inglés) para el RNA viral y la determinación de una respuesta inmune anti-PRRSV, utilizando una nueva ELISA para medir las respuestas de IgG específicas de perritos de la pradera a nueve polipéptidos virales. Resultados: Una variedad de tejidos resultaron negativos al RNA del PRRSV durante todo el estudio tanto en animales tratados como en los controles. Los signos clínicos fueron poco remarcables y no se observaron lesiones histopatológicas del PRRS. No se observó seroconversión en ningún animal durante el estudio de 28 días. Se observaron variaciones individuales en los niveles de fondo de anticuerpos y resultados de RT-PCR. Implicaciones: Los perritos de la pradera no permiten la replicación del PRRSV y no son un reservorio del virus. Los valores cuantitativos de RT-PCR para PRRSV en las muestras de suero y de tejido se traslapan con valores de fondo negativos cerca de los límites de detección, aumentando el riesgo de interpretaciones falsas positivas. Los métodos, materiales, y recursos para la investigación diagnóstica del perrito de la pradera ya están disponibles para la comunidad veterinaria. | ResuméObjectif: Déterminer si le chien de prairie est un hôte biologique pour le virus du syndrome reproducteur et respiratoire porcin (PRRSV). Matériels et méthodes: Seize chiens de prairie capturés en nature ont été inoculés par voie intrapéritonéale avec un mélange à titre élevé de souches sauvages locales du PRRSV, et sept animaux ont servi de témoins non-inoculés. Du sérum et des tissus ont été prélevés à des intervalles de 3 à 7 jours pendant 28 jours pour analyse par réaction d’amplification en chaîne par la polymérase avec la transcriptase inverse (RT-PCR) de l’ARN viral et détermination d’une réponse immune anti-PRRSV à l’aide d’une nouvelle épreuve ELISA mesurant la réponse IgG spécifique du chien de prairie à neuf polypeptides viraux. Résultats: Plusieurs tissus se sont avérés négatifs pour la présence d’ARN du PRRSV à tous les prélèvements autant chez les animaux traités que les témoins. Les signes cliniques étaient non-significatifs et aucune lésion histopathologique de PRRSV n’a été notée. Aucune séroconversion n’a été observée chez les animaux durant la période de 28 jours de l’étude. Des variations individuelles des niveaux de base des anticorps et des résultats de RT-PCR ont été observées. Implications: La réplication du PRRSV ne se produit pas chez les chiens de prairie et ces derniers ne sont donc pas des réservoirs du virus. Les valeurs quantitatives de RT-PCR pour le PRRSV dans le sérum et les échantillons de tissu chevauchent les valeurs négatives de bruit de fond près des limites de détection, augmentant ainsi le risque d’interprétation de faux-positifs. Des méthodes, du matériel et des ressources pour des enquêtes diagnostiques chez les chiens de prairie sont maintenant accessibles à la communauté vétérinaire. |
Keywords: swine, biosecurity,
porcine reproductive and respiratory syndrome virus, PRRSV, polymerase chain
reaction, PCR, wildlife
Search the AASV web site
for pages with similar keywords.
Received: February
15, 2006
Accepted: April
6, 2006
Transmission of porcine reproductive and respiratory syndrome virus (PRRSV) among farms may occur by a variety of means in spite of extensive biosecurity safeguards. The role of wildlife reservoirs in PRRSV transmission has not been demonstrated, outside of one observation of shedding in ducks that has not been replicated.1,2 Nevertheless, wildlife species are often reservoirs of animal disease, and they might provide a natural reservoir for PRRSV. Here, we report on an investigation of the prairie dog (Cynomys ludovicianus), a rodent species, as a possible reservoir of PRRSV. Prairie dogs are widely distributed in the central and southwestern United States, forming large colonies containing hundreds of individuals. We reasoned that rodent species such as the prairie dog are potential reservoirs of PRRSV, since the house mouse, Mus musculus, is the natural host for lactate dehydrogenase elevating virus, the arterivirus most closely related to PRRSV.3
The possibility that prairie dogs might circulate PRRSV in a local area was raised by a series of outbreaks in a breeding swine herd in the southwestern United States. Diagnostic ORF 5 sequencing has shown that PRRSV genotypes that most closely matched isolates from the past reappeared periodically over a 5-year period, yet no new viruses appeared to have been introduced from breeding stock replacements, semen, or area spread in this large and isolated production system.
The objective of this study was to determine if prairie dogs could be a wildlife reservoir of PRRSV. For a period of 28 days after inoculation of captive wild prairie dogs with PRRSV, serological testing for antibody formation, and sequential virological and histopathological examination of tissues for evidence of viral RNA and classical lesions of PRRS were performed.
Materials and methods
Animals and housing
A total of 23 male and female prairie dogs of unknown age were captured by live trapping for approximately 3 months from a wild colony in the midst of a swine operation in the southwestern United States, located more than 80 km from the nearest other swine operation. Animal capture and care procedures were approved by the Food and Drug Administration Center for Veterinary Medicine. Animals were housed by treatment group (controls and infected) at CAVL Diagnostics (Amarillo, Texas) in two rooms with separate ventilation systems. Animals were provided with timothy hay (Kaytee Products, Inc, Chilton, Wisconsin) and water ad libitum, and were limit-fed rodent chow (Bluebonnet Feeds, Brownwood, Texas). Controls were always handled first. Footwear, outerwear, and gloves were changed between groups.
Study design
Serum samples collected from all animals prior to the experiment tested negative for PRRSV by quantitative real-time reverse transcriptase-polymerase chain reaction (RT-PCR) and for antibody by enzyme-linked immunosorbent assay (ELISA).
Animals were randomly assigned to the control group (n = three females and four males) and the treatment group (n = seven females and nine males). On Day 0, animals in both groups were anesthetized with a reconstituted combination of tiletamine HCl and zolazepam HCl (Telazol; Fort Dodge Animal Health, Fort Dodge, Iowa) at 0.55 mL per kg body weight injected intramuscularly. Treatment animals were then inoculated intraperitoneally with a high-titered mixture of six wild-type PRRSV strains. Control animals were not inoculated.
As it was possible that PRRSV might reside in tissues from animals showing little or no viremia (compared to pigs), two inoculated animals and one control were sacrificed on each of Days 0, 3, 7, 10, 14, 21, and 28, and quantitative RT-PCR for PRRSV was performed on tissues from lung, heart, liver, kidney, spleen, thymus, tonsil, salivary gland, and pulmonary and mesenteric lymph nodes (ie, tissues from which PRRSV is commonly isolated in swine). Animals were sacrificed by inducing anesthesia as described, then exsanguinating via cardiac puncture. Tissues were also harvested from an inoculated prairie dog that died on Day 8 due to an anesthesia complication, and from an additional treatment-group prairie dog on Day 28. One hundred to 700 mg of tonsil, pulmonary lymph node, mesenteric lymph node, and lung tissue were preserved in 5 mL of RNAlater (Ambion, Austin, Texas) and stored at 4°C for quantitative RT-PCR. The remaining tissues were stored in formalin at 4°C.
Blood samples were collected by cardiac puncture from all animals remaining at each time point and frozen at -20°C for ELISA and quantitative RT-PCR. Blood collection was facilitated by Telazol anesthesia as described. The amount of serum obtained from blood samples varied between 500 and 1000 µL per animal.
Viruses
Six PRRSV isolates from infected pigs in the swine operation
were grown on MA-104 cells or MARC 145 cells to the following
titers (in median tissue culture infective doses per mL; TCID50 per
mL): 04-45228-31, 5 × 104; 04-45229-33, 5 ×
106; SD PRRSV 04-100, 5 × 103; SD PRRSV
04-101, 5 × 102; SD PRRSV 04-102,
5 × 103; and SD PRRSV 04-103, 5 × 102. The
first two strains were kindly provided by Dr Marie Gramer,
University of Minnesota Veterinary Diagnostic Laboratory, St Paul,
Minnesota, and the last four by Dr Jane Christopher-Hennings,
Animal Disease Research and Diagnostic Laboratory, South Dakota
State University, Brookings, South Dakota. Equal aliquots were
combined to make a 6-mL inoculum in cell culture medium that
contained 1 mL of each viral isolate.
Histopathology
Samples of lung, pulmonary lymph node, spleen, and heart from control and inoculated animals sacrificed on Days 0, 7, and 21 were submitted in formalin to the Minnesota Veterinary Diagnostic Laboratory.
RNA isolation and quantitative RT-PCR
Isolation of RNA was performed for all samples using a Nucleospin II RNA Isolation kit (BD Biosciences, San Jose, California). A titered PRRSV strain, MN30100, was aliquoted into a series of eight 10-fold dilutions that were used to generate a positive-control RT-PCR standard curve.4 RNA was extracted from 100 µL of each dilution, eluted in 50 µL RNase-free water provided in the kit, and stored at -80°C. For prairie dog serum samples, RNA was extracted from 150 to 200 µL of serum, eluted in 50 µL of RNase-free water, and stored at -80°C. Tissue samples were weighed and homogenized in five volumes of lysis buffer using a Polytron PT 3100 (Brinkman, Westbury, New York). Two hundred µL of homogenate was extracted, and purified RNA was eluted in 50 µL RNase-free water for storage at -80°C. For real-time RT-PCR assays, samples were dried in a Savant SpeedVac (Thermo Electron Corp, Waltham, Massachusetts) and resuspended in 5 µL of RNase-free water.
Primers and dual-labeled FAM-TAMRA probe were designed from the ORF 7-3’UTR region of PRRSV.5 Real-time RT-PCR was carried out with an ABI 7500 Sequence Detection System (Applied Biosystems, Foster City, California). PRRS virus RNA reverse transcription and PCR were performed on duplicate samples in a single one-step TaqMan reaction mix (Applied Biosystems). Each 20-µL reaction contained 10.0 µL TaqMan One-Step Mastermix, 0.2 µL enzyme mix (catalog no. 4309169; Applied Biosystems), 0.3 µM forward primer (5’ tgatgggctggcattctt 3’), 0.3 µM reverse primer (5’ acacggtcgccctaattg 3’), 0.2 µM dual-labeled probe (6-FAM- tgtggtgaatggcactgattgaca-TAMRA), 2 µL extracted RNA, and 5.6 µL RNase-free water. Thermocycler conditions were 50°C for 30 minutes followed by 95°C for 10 minutes, then 45 cycles of 95°C for 15 seconds and 60°C for 1 minute. Data were recorded as the cycle at which the amount of fluorescence exceeded an arbitrary threshold (Ct). Reactions that did not cross the threshold at the end of 45 cycles were assigned a Ct value of 45.
ELISA for PRRSV-specific antibodies in prairie dog serum
As seroconversion in prairie dogs might be a more sensitive indicator of PRRSV infection at low levels or in tissues not tested by RT-PCR, an ELISA test was developed to detect antigen-specific humoral immune responses to PRRSV. Nine recombinant PRRSV proteins from strain VR2332 were expressed from the plasmid pET 24b as fusion proteins containing an amino terminal myc-tag and a carboxyl terminal 6× histidine tag in BL 21(DE3)-RP cells (Stratagene, La Jolla, California) (Table 1). For purification, denatured proteins were dialyzed overnight in 0.1 M Tris HCl (pH 8.0), 6 M guanidine HCl, and 2 mM EDTA at 4°C, then adjusted to a concentration of 3 mg per mL. Dithiothreitol was added to a final concentration of 300 mM and the solution was filtered through a 0.45-µm membrane. Reduced protein was added into refolding buffer (100 mM Tris HCl (pH 8.0); 0.5 M L-arginine; 8 mM oxidized glutathione; 2 mM EDTA; 10 µM pepstatin A; 10 µM leupeptin; and 1 mM phenylmethylsulfonylflouride), filtered (0.22 µm), and stirred overnight at 4°C.6,7 The purified protein was concentrated by tangential flow filtration (Pellicon XL Ultracel PLC 5 kd; Millipore, Billerica, Massachusetts) and dialyzed against 20 mM Tris HCl (pH 8.0) and 150 mM NaCl. Protein concentration was determined by Bradford assay (BioRad, Hercules, California). Purified protein solutions were stored at -80°C before coating on ELISA plates.
Table 1: Porcine reproductive and respiratory syndrome virus (PRRSV) proteins and polypeptides used in an ELISA for testing serum of wild-caught prairie dogs inoculated intraperitoneally with six strains of PRRSV
* Nucleotide sequence ranges are from Genbank accession #U87392. |
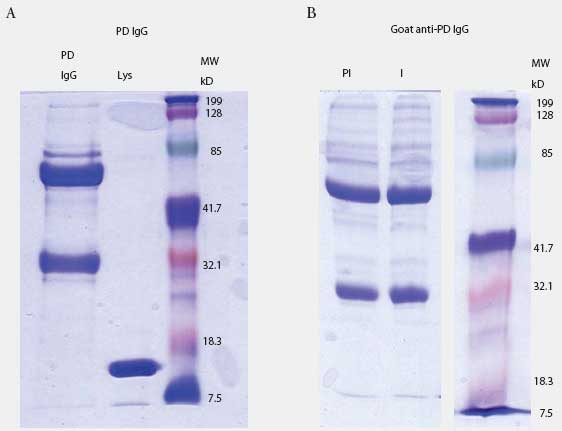
Prairie dog IgG-specific antisera were produced in goats. The IgG fraction was isolated from pooled prairie dog serum (collected from five animals before the experiment started) by three precipitations with 40% saturated ammonium sulfate. Concentration of IgG was determined by spectrophotometry (Nanodrop ND-100; Nanodrop Technologies, Wilmington, Delaware) using an extinction coefficient E1%280 = 13.7. Purity was determined by 10% SDS-PAGE analysis (Figure 1A). Purified material was used to immunize goats to produce anti-prairie dog IgG antiserum (Bethyl Laboratories, Montgomery, Texas). Goat IgG containing antibodies to prairie dog IgG was purified by triple 40% saturated ammonium sulfate precipitation, dialysis in phosphate buffered saline (PBS), and adjustment to the original serum volume (Figure 1B).
| Figure 1: Prairie dog (PD) IgG purification and
anti-IgG production in goat. Figure 1A: prairie dog IgG was purified by
multiple rounds of ammonium sulfate precipitation and visualized by SDS-PAGE.
Lane 1: 10 µg purified IgG; Lane 2: lysozyme (Lys); Lane 3: molecular
weight (MW) standards. Figure 1B: goat anti-PD IgG purified by ammonium
sulfate precipitation before (lane PI) and after (lane I) immunization
with purified PD IgG. Each lane contains 5 µg of total protein. Lane
3 contains MW markers.
|
ELISA for PRRSV-specific antibodies was performed by coating microtiter plates with 300 ng of each of the nine recombinant PRRSV proteins in carbonate buffer (pH 9.6), blocking with 5% nonfat dry milk in PBS containing 0.05% Tween 20, and reacting with serum samples diluted 1:100 in PBS containing 5% nonfat dry milk and 0.05% Tween-20. Wells were washed and replicate samples were incubated with 100 µL of pre-immune or immune goat anti-prairie dog IgG diluted 1:100. After 1 hour, the plates were washed and developed with horseradish peroxidase-conjugated rabbit anti-goat IgG in a dilution of 1:5000 for 1 hour. Plates were then washed and color was developed with tetramethyl blue tetrazolium substrate (KPL, Gaithersburg, Maryland). Reactions were stopped with 1 M phosphoric acid and plates were read at 450 nm.
ELISA for prairie dog IgG was performed by coating microtiter plates with 0 to 320 ng of prairie dog IgG per well as described above. Blocking and washing steps were performed as described above. Samples were incubated with 100 µl of pre-immune or immune IgG in 10-fold dilutions from 1:100 to 1:107 for 1 hour. Color development was performed as described above, using horseradish peroxidase-conjugated rabbit anti-goat IgG to detect the goat anti-prairie dog IgG bound to prairie dog IgG.
Since the ELISA involves three binding reactions, we were concerned that negative results might indicate a failure of the assay to detect prairie dog antibody binding to plate-bound antigen. To address this possibility, we determined the ability of prairie dog natural antibodies to react with bacterial antigens instead of PRRSV antigens on an ELISA plate. ELISA for prairie dog natural antibodies was performed by preparing an overnight culture of DH5a Escherichia coli, treating with lysozyme for 30 minutes, heating at 90°C for 5 minutes, and pelleting at 10,000g for 10 minutes. The pellet was resuspended in one-tenth volume of PBS, sonicated, and coated undiluted and at ten-fold dilutions in 200 µL total volume at pH 9.4, as described above. Escherichia coli antigens were detected with three antibody reactions, consisting of prairie dog IgG followed by goat anti-prairie dog IgG and horseradish peroxidase-conjugated rabbit anti-goat IgG. Prairie dog sera from one control and two PRRSV-inoculated animals at Day 28 were diluted 1:100 and applied in duplicate to the wells. Blocking and washing steps were performed as described above. All wells were then treated with a 1:100 dilution of goat pre-immune IgG or immune anti-prairie dog IgG. Immune complexes of prairie dog IgG and goat anti-prairie dog IgG were detected with horseradish peroxidase-conjugated rabbit anti-goat IgG as described above.
Results
Histopathology
In prairie dogs sacrificed on Day 0 (before inoculation with PRRSV), heart, lung, spleen, and pulmonary lymph node were devoid of lesions that could be attributable to PRRSV infection. One animal had evidence of myocardial inflammation which had been resolved by fibrosis. Partial cuffing by small lymphoid nodules was noted in lungs of two animals and local replacement of myocardial tissue with fibrous connective tissue was noted in one animal. The same observations were made on Day 7, with additional observation of mild cardiac thickening due to subacute inflammation or infiltration by lymphocytes and macrophages. Histopathology was unchanged in control and inoculated animals at Day 21 except for clear cytoplasmic vacuoles in a few alveolar macrophages of the control animal, and focal inflammation centered around inhaled plant material in one inoculated animal. Overall, no signs of PRRS, including microscopic interstitial pneumonia and cardiac arteritis, were noted in any animals.
RT-PCR screening for PRRSV
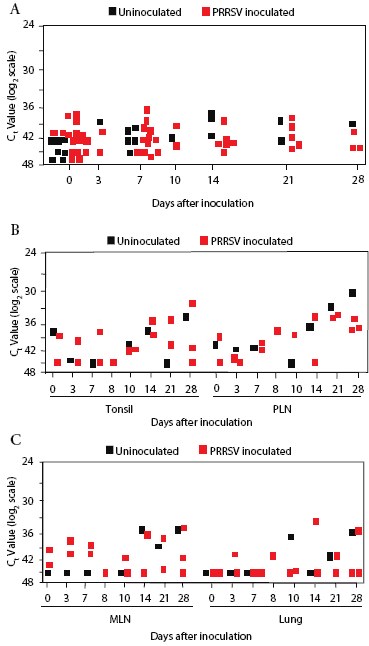
Background real-time RT-PCR threshold cycle (Ct) values for prairie dog serum, determined on samples from all animals before inoculation, were in the range of 36 to 45 (Figure 2). Sample Ct values remained within this range at all sampling times after inoculation through Day 28. There was no difference in values in serum from inoculated and control prairie dogs (Figure 2).
| Figure 2: Quantitative real-time reverse transcriptase-polymerase
chain reaction screening for PRRSV RNA in prairie dog serum and tissue
samples. Prairie dogs were inoculated intraperitoneally with PRRSV (Treated;
n = 16) or not inoculated (Control; n = 7) on Day 0 and were sacrificed
for collection of tissue samples between Days 0 and 28. RNA was extracted
from 200 µL of serum (Figure 2A) or 200 mg of tissue (Figures 2B
and 2C) and processed for amplification of PRRSV RNA. Data were recorded
as the cycle at which the amount of fluorescence exceeded an arbitrary
threshold (Ct). The results show the obtained Ct values on a log2 scale.
Reactions that did not cross the threshold at the end of 45 cycles were
assigned a Ct value of 45. Each symbol represents the average of duplicate
tests from one animal. PLN, peripheral lymph node; MLN, mesenteric lymph
node.
|
The overall Ct range was 29 to 45 in tonsil, pulmonary lymph node, mesenteric lymph node, and lung samples collected on Days 0, 3, 7, 10, 14, 21, and 28 (Figure 2). Moreover, Ct values for tissues from control animals were frequently lower than for tissues from inoculated prairie dogs, further indicating that PRRSV RNA was not present, even though there was a wide range of results (Figure 2).
Anti-PRRSV antibody responses
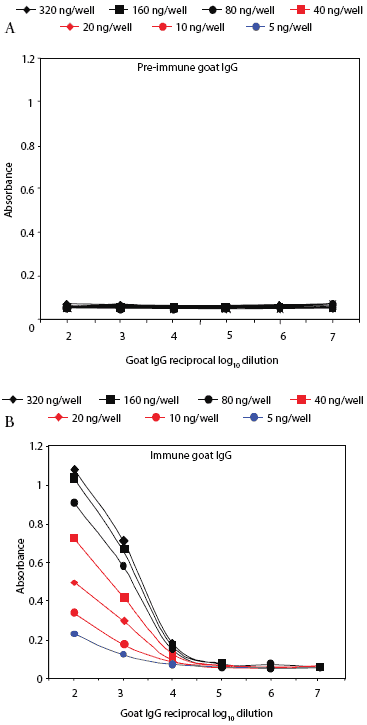
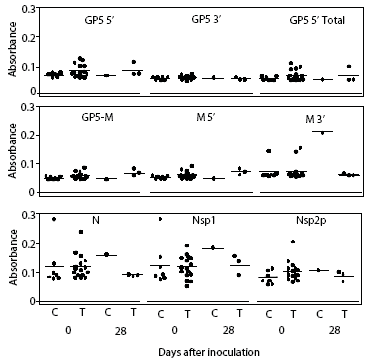
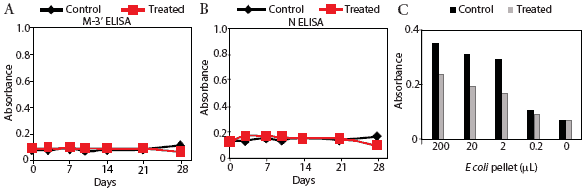
Prairie dog serum contained approximately 12.6 ± 6.1 mg IgG per mL (mean ± SE). A checkerboard titration (ie, varying amounts of prairie dog IgG coated on the plates and varying amounts of pre-immune or immune goat antisera tested) was performed to determine the conditions for assessing the amount of anti-PRRSV antibody response. Prairie dog IgG-specific, dose-dependent immunoreactivity was readily detected (Figure 3). No PRRSV antigen-specific reactivity was observed against any of the nine PRRSV proteins or protein fragments in the 23 animals on Day 0 (ie, they had no pre-existing immunity to PRRSV). At 28 days post infection, sera were similarly negative for all PRRSV antigens, albeit with sporadic high absorbance values (Figure 4). Because only three infected animals remained at 28 days, serum samples were evaluated further for serological response to N and M-3’ antigens at all time points following infection. As shown in Figures 5A and 5B, individual absorbance values were the same in control and test animals, indicating the absence of anti-PRRSV antibodies.
| Figure 3: Specific IgG antibody response of a goat
immunized with prairie dog IgG. Reactivity of pre-immune (Figure 3A) and
immune (Figure 3B) goat IgG is shown against prairie dog IgG coated in
varying amounts (5-320 ng per well) onto microtiter plates. Uncoated wells
gave absorbance values of ≤ 0.06.
|
| Figure 4: Lack of anti-PRRSV antibodies in sera
from prairie dogs inoculated intraperitoneally with PRRSV on Day 0. Day
0 serum samples from uninoculated controls (C; n = 7) and inoculated animals
(T; n = 16) and Day 28 samples from surviving animals (C, n = 1; T, n =
3) were diluted 1:100 and applied to plates coated with 100 ng of each
of nine different PRRSV proteins or polypeptide constructs (Table 1). Unadjusted
OD values for each animal are shown at both days for each protein.
|
| Figure 5: Absence of specific anti-PRRSV antibody
response in prairie dogs. Prairie dog serum samples were obtained from
0 to 28 days after animals were inoculated with PRRSV (Treated) or not
inoculated (Control). Serum was applied to microtiter plates coated with
membrane protein (M-3’; Figure 5A) or nucleocapsid (N; Figure 5B),
reacted with goat anti-prairie dog IgG, and detected with horseradish peroxidase-conjugated
rabbit anti-goat IgG. Figure 5C: Detection of Control (n = 1) and Treated
(n = 2) serum reactivity to bacterial lysate antigens. Plates were coated
with various dilutions of Escherichia coli lysate and reacted with
Day 28 prairie dog IgG. Absorbance was determined following incubation
with immune goat anti-prairie dog IgG, horseradish peroxidase-conjugated
rabbit anti-goat IgG, and color substrate.
|
Results of the E coli ELISA for prairie dog natural antibodies are shown in Figure 5C. Specific reactivity was observed only in wells containing control and inoculated prairie dog IgG and goat anti-prairie dog IgG. Pre-immune goat IgG values were the same as the negative control that included no antigen (data not shown). This result shows that the method involving three antibody-binding reactions detected prairie dog antibodies and supports the conclusion that prairie dogs did not produce an antibody response to PRRSV inoculation.
Discussion
Starting in 1998, PRRSV ORF 5 sequence information was collected in the isolated swine production system where chronic PRRS re-infection problems raised the original questions about prairie dogs as a potential zoonotic reservoir of PRRSV. After the year 2000, the data, consisting of > 600 sequences, indicated that no new viruses had entered the system from external, lateral introductions. Instead, viral isolates from periodic episodes of abortion and nursery mortality spikes appeared more similar to historical isolates than to contemporaneously occurring forms in the system. Many of these sequences differed by as much as 4.5% from circulating strains, but differed from older isolates by less than 2% on sequence analysis.
An explanation for this phenomenon could be that the large multi-site complex acted as a geographic continuous-flow system, and the less dominant older virus types remained present at low levels in the growing-pig units. For this to be the case, the virus would have to be efficiently spread by aerosol, infected-pig movement, or an unknown mechanical vector. Aerosol spread appears unlikely, since the re-appearance of older isolates followed no apparent pattern. Pig movement could not be linked to re-breaks. Biosecurity at the time of the study was far more stringent than in other commercial systems in the United States. The system is in an isolated area more than 80 km from other pig production units. Transport vehicles were pressure-washed with heated water and dried for 24 hours. Personnel were dedicated to specific parts of the system, 48- to 96-hour downtime rules were enforced, and the site was fenced and patrolled continuously by security guards. The boar stud remained PRRSV-negative during the study period. Outside breeding stock (boars and gilts) were from known negative sources and remained negative until exposure after arrival.
The strict biosecurity measures and results of viral sequencing analysis seemed to preclude external sources of new infections. Thus, it was possible that older PRRSV isolates were being maintained in a nonporcine rodent reservoir such as the indigenous prairie dog. However, the immunological, virological, histopathological, and clinical findings presented here rule out the prairie dog as a host species for PRRSV. The source of PRRSV before its discovery as a viral pathogen of swine in the 1980s remains to be determined.8,9 Mice were determined previously not to be a host species for PRRSV,10 and a report of PRRSV growth and shedding in ducks could not be reproduced.2 Until the host range possibilities of PRRSV are more fully investigated, the evolutionary trajectory and origins of PRRSV, including the hypothesis of a non-swine reservoir, are speculative.11,12 The findings help to focus attention on mechanisms of viral transmission by excluding the prairie dog as a possible reservoir and vector.
Screening of serum and tissues by RT-PCR for viral RNA revealed a wide range of Ct values for negative samples. Real-time PCR is a powerful method for detection and quantification of nucleic acids, but its application to diagnostic specimens gives results that may show overlap between weakly positive samples and negative samples with high backgrounds. Here, serum samples from negative animals gave Ct values as low as 36 cycles, and tissue samples gave Ct values as low as 29 cycles, which is well within the range of true-positive results.13 These findings emphasize the reality that tissue samples, in particular, are subject to high backgrounds and spurious false-positive results, and that RT-PCR should not be relied on exclusively to provide evidence of PRRSV infection, especially when additional test methods, such as serology, are available, or when the PCR results are near the limit of assay sensitivity.
New information also was gained about humoral IgG of the prairie dog. The concentration of IgG in serum of wild-caught adults, 12.6 ± 6.1 mg per mL, is less than the serum IgG concentration in swine (20 to 30 mg per mL, unpublished data, W. Yu and M. P. Murtaugh, 2005), but is similar to that in humans (normal range = 8 to 16 mg per mL14,15). While the prairie dog is not a reservoir for PRRSV, it is a reservoir of human disease.16-18 Thus, it might play a role in the spread of other swine diseases. The methods and reagents presented here would then be useful for development of appropriate diagnostics.
Implications
- The prairie dog can be excluded as a PRRSV reservoir in unexplained PRRS outbreaks in the geographic home range of the animal.
- Methods, materials, and resources for diagnostic investigation of the prairie dog are now publicly available to the swine veterinary community.
- More detailed information about the sensitivity and specificity of real-time RT-PCR assays for PRRSV RNA in serum and tissue samples would help in diagnostic interpretations.
Acknowledgements
We thank Dr Richard Mock at the Texas Veterinary Medical Diagnostic Laboratory, Amarillo, Texas, for assistance in identifying resources familiar with capture, housing, and care of prairie dogs, and Dr Marie Gramer, Minnesota Veterinary Diagnostic Laboratory, St Paul, and Dr Jane Christopher-Hennings, South Dakota Animal Disease Research and Diagnostic Laboratory, Brookings, for providing virus samples. Colleen Finnegan, University of Minnesota, provided expert technical support. Partial funding support for the research was provided by Boehringer Ingelheim Vetmedica. Dr Dale Polson is an employee of Boehringer Ingelheim Vetmedica.
References
1. Zimmerman JJ, Yoon KJ, Pirtle EC, Wills RW, Sanderson TJ, McGinley MJ. Studies of porcine reproductive and respiratory syndrome (PRRS) virus infection in avian species. Vet Microbiol. 1997;55:329–336.
2. Trincado C, Dee S, Rossow K, Halvorson D, Pijoan C. Evaluation of the role of mallard ducks as vectors of porcine reproductive and respiratory syndrome virus. Vet Rec. 2004;154:233–237.
3. Plagemann PGW. Lactate dehydrogenase-elevating virus and related viruses. In: Fields BN, Knipe DM, Howley PM, eds. Fields Virology. 3rd edition. Philadelphia: Lippincott-Raven; 1996:1105–1120.
4. Bierk MD, Dee SA, Rossow KD, Otake S, Collins JE, Molitor TW. Transmission of PRRS virus from persistently infected sows to contact controls. Can J Vet Res. 2001;65:261–266.
5. Schurrer JA, Dee SA, Moon RD, Murtaugh MP, Finnegan CP, Deen J, Kleiboeker SB, Pijoan C. Retention of ingested porcine reproductive and respiratory syndrome virus in house flies. Amer J Vet Res. 2005;66:1517–1525.
6. Buchner J, Pastan I, Brinkmann U. A method for increasing the yield of properly folded recombinant fusion proteins: single-chain immunotoxins from renaturation of bacterial inclusion bodies. Anal Biochem. 1992;205:263–270.
7. Clark EDB. Refolding of recombinant proteins. Curr Opin Biotechnol. 1998;9:157–163.
8. Wensvoort G, Terpstra C, Pol JMA, ter Laak EA, Bloemraad M, de Kluyver EP, Kragten C, van Buiten L, den Besten A, Wagenaar F, Broekhuijsen JM, Moonen PLJM, Zetstra T, de Boer EA, Tibben HJ, de Jong MF, van’t Veld P, Groenland GJR, van Gennep JA, Voets MT, Verheijden JHM, Braamskamp J. Mystery swine disease in the Netherlands: The isolation of Lelystad virus. Vet Q. 1991;13:121–130.
9. Carman S, Sanford SE, Dea S. Assessment of seropositivity to porcine reproductive and respiratory syndrome (PRRS) virus in swine herds in Ontario – 1978 to 1982. Can Vet J. 1995;36:776–777.
10. Hooper CC, Van Alstine WG, Stevenson GW, Kanitz CL. Mice and rats (laboratory and feral) are not a reservoir for PRRS virus. J Vet Diagn Invest. 1994;6:13–15.
11. Plagemann PG. Porcine reproductive and respiratory syndrome virus: origin hypothesis. Emerg Infect Dis. 2003;9:903–908.
12. Hanada K, Suzuki Y, Nakane T, Hirose O, Gojobori T. The origin and evolution of porcine reproductive and respiratory syndrome viruses. Mol Biol Evol. 2005;22:1024–1031.
13. Wasilk A, Callahan JD, Christopher-Hennings J, Gay TA, Fang Y, Dammen M, Reos ME, Torremorell M, Polson D, Mellencamp M, Nelson E, Nelson WM. Detection of U.S., Lelystad, and European-like porcine reproductive and respiratory syndrome viruses and relative quantitation in boar semen and serum samples by real-time PCR. J Clin Microbiol. 2004;42:4453–4461.
14. Hamilton RG. The Human IgG Subclasses. La Jolla, California: Calbiochem-Novabiochem; 1998:28.
15. Kuby J. Immunology. New York: WH Freeman and Company; 1996:124–126.
16. Cully JF Jr, Carter LG, Gage KL. New records of sylvatic plague in Kansas. J Wildl Dis. 2000;36:389–392.
17. Petersen JM, Schriefer ME, Carter LG, Zhou Y, Sealy T, Bawiec D, Yockey B, Urich S, Zeidner NS, Avashia S, Kool JL, Buck J, Lindley C, Celeda L, Monteneiri JA, Gage KL, Chu MC. Laboratory analysis of tularemia in wild-trapped, commercially traded prairie dogs. Emerg Infect Dis. 2004;10:419–425.
18. Xiao SY, Sbrana E, Watts DM, Siirin M, da Rosa AP, Tesh RB. Experimental infection of prairie dogs with monkeypox virus. Emerg Infect Dis. 2005;11:539–545.