Case study |
Peer reviewed |
Tonsilar crypt exudate to evaluate shedding and transmission of porcine reproductive and respiratory syndrome virus after inoculation with live field virus or vaccination with modified live virus vaccine
Uso de exudado de las criptas de las amígdalas de cerdos vivos para evaluar la eliminación y la transmisión del virus del síndrome reproductivo y respiratorio porcino después de la inoculación con virus vivo de campo o vacunados con vacuna de virus vivo modificada
Utilisation d’un exsudat des cryptes tonsillaires provenant de porcs vivants pour évaluer l’excrétion et la transmission du virus du syndrome reproducteur et respiratoire porcin après inoculation avec un virus de champs vivant ou vaccination avec un vaccin à virus vivant modifié
Thomas J. Fangman, DVM, MS, Diplomate ABVP-SHM; Steve B. Kleiboeker, DVM, PhD; Melinda Coleman, DVM
TJF: University of Missouri, College of Veterinary Medicine, University of Missouri, Columbia, Missouri. SBK: Viracor, Lee Summit, Missouri. MC: Albany Veterinary Services, Albany, Missouri. Corresponding author: Dr Thomas J. Fangman, University of Missouri, A331 Clydesdale Hall, Columbia, MO 65211; Tel: 573-882-788; Fax: 573-884-9139; E-mail: fangmant@missouri.edu.
Cite as: Fangman TJ, Kleiboeker SB, Coleman M. Tonsilar crypt exudate to evaluate shedding and transmission of porcine reproductive and respiratory syndrome virus after inoculation with live field virus or vaccination with modified live virus vaccine. J Swine Health Prod. 2007;15(4):219–223..
Also available as a PDF.
SummaryTwo isolated groups of 44 seven-week-old principal pigs were confirmed negative for porcine reproductive and respiratory syndrome virus (PRRSV) by testing blood samples by ELISA and reverse transcription-polymerase chain reaction (RT-PCR). Pigs were then inoculated (Day 0) with serum containing a field strain of PRRSV (LVI) or were vaccinated with a commercial modified live virus PRRS vaccine (MLV). Blood samples and tonsilar scrapings were collected on Day 14 and tonsilar scrapings on Days 28, 42, 56, 70, 100, 130, and 160 for testing by RT-PCR soon after collection. When 90% of pigs were PCR-negative (Day 130), three PRRSV-naive sentinel pigs were introduced per pen of LVI and MLV pigs. No sentinels had seroconverted when tested by ELISA 30 days later. Tonsilar crypt exudate and RT-PCR-positive serum samples from the principal pigs were pooled to create homogenates for a swine bioassay. Three-week-old PRRSV-naive pigs were inoculated intramuscularly with PRRSV-positive tonsilar crypt exudate pools (15 pigs), PRRSV-positive serum pools (five pigs), or PRRSV-negative tonsilar crypt exudate pools (five pigs). When tested by ELISA 30 days later, all 20 pigs inoculated with PRRSV-positive pools were seropositive and the five pigs inoculated with PRRSV-negative pools were seronegative. | ResumenSe confirmó que dos grupos de 44 cerdos experimentales de siete semanas de edad eran negativos al virus del síndrome reproductivo y respiratorio porcino (PRRSV por sus siglas en inglés) mediante muestra de sangre probadas con ELISA y la prueba de transcriptasa reversa de la reacción en cadena de la polimerasa de (RT-PCR por sus siglas en inglés). Después se inocularon los cerdos (Día 0) con suero que contenía una cepa de campo del PRRSV (LVI) o fueron vacunados con una vacuna viva comercial del virus vivo modificado de PRRS (MLV por sus siglas en inglés). Se recolectaron muestras de sangre y raspados de amígdalas en el Día 14 y raspados de amígdalas los Días 28, 42, 56, 70, 100, 130, y 160 para ser probadas mediante RT-PCR poco tiempo después de la recolección. Cuando el 90% de los cerdos fueron negativos al PCR (Día 130), se introdujeron tres cerdos centinela, libres de PRRSV por cada corral de cerdos LVI y MLV. Ningún centinela seroconvirtió cuando se probaron por la prueba de ELISA 30 días después. El exudado de cripta y las muestras de suero positivas a RT-PCR de los cerdos experimentales se mezclaron para crear homogeneizado para bioensayo porcino. Se inocularon intramuscularmente cerdos de tres semanas de edad libres de PRRSV con pools de muestras positivas al PRRSV del exudado de cripta de amígdalas (15 cerdos), pools de sueros positivos al PRRSV (cinco cerdos), o pools de exudado de cripta de amígdalas negativo al PRRSV (cinco cerdos). Cuando se probaron por ELISA 30 días después, los 20 cerdos inoculados con pools positivos al PRRSV resultaron seropositivos y los cinco cerdos inoculados con pools negativos al PRRSV fueron seronegativos. | ResuméDeux groupes isolés de 44 porcs âgés de 7 semaines (groupes principaux) ont été confirmés négatifs pour la présence du virus du syndrome reproducteur et respiratoire porcin (PRRSV) dans des échantillons de sang par épreuve ELISA et par épreuve d’amplification en chaîne par la polymérase à l’aide de la transcriptase réverse (RT-PCR). Les porcs ont ensuite été inoculés (Jour 0) avec du sérum contenant un isolat de champs de PRRSV (LVI) ou ont été vaccinés avec un vaccin commercial contenant un virus vivant modifié du PRRS (MLV). Des échantillons sanguins et des grattages des amygdales ont été prélevés au Jour 14 et des grattages des amygdales prélevés aux Jours 28, 42, 56, 70, 100, 130, et 160 pour être éprouvés par RT-PCR peu de temps après leur collecte. Lorsque 90% des porcs se sont révélés négatifs par PCR (Jour 130), trois porcs sentinelles naïfs pour le PRRSV ont été introduits par parc de porcs LVI et MLV. Aucun des animaux témoins n’avait développé d’anticorps lorsque testé 30 jours plus tard par ELISA. Les exsudats des cryptes tonsillaires et les échantillons de sérum positifs par RT-PCR provenant des groupes de porcs principaux ont été regroupés pour former des homogénats devant servir dans un bio-essai chez les porcs. Des porcs naïfs âgés de 3 semaines ont été inoculés par voie intramusculaire avec un pool d’exsudats de cryptes tonsillaires PRRSV-positif (15 porcs), un pool d’échantillons de sérum PRRSV-positif (cinq porcs), ou un pool d’exsudats de cryptes tonsillaires PRRSV-négatif (cinq porcs). Lorsque testé 30 jours plus tard par ELISA, les 20 animaux inoculés avec les pools PRRSV-positifs se sont révélés séropositifs et les cinq porcs inoculés avec les pools PRRSV-négatifs sont demeurés séronégatifs. |
Keywords: swine, porcine
reproductive and respiratory syndrome virus, PRRSV, persistent infection,
tonsil
Search the AASV web site
for pages with similar keywords.
Received: January
8, 2007
Accepted: February
7, 2007
Porcine reproductive and respiratory syndrome (PRRS) is an important disease of swine, causing viremia, severe and sometimes fatal respiratory and reproductive disease, and significant production losses. The PRRS virus (PRRSV)1 is one of the primary agents contributing to porcine respiratory disease complex (PRDC), another economically important disease of swine. Many pigs make a full clinical recovery from PRRS, yet carry a low-level viral infection for an extended period. Persistent infection with PRRSV has been well documented under experimental conditions.2–7 “Carrier” pigs shed the virus either intermittently or continuously, infecting naive pigs by direct or indirect contact. Persistence of PRRSV infection in individual pigs is one of the major impediments to PRRS control.
Results of our previous study8 suggest that persistently infected pigs can be identified if an appropriate sample type and detection method are used. Under experimental conditions, oropharyngeal scrapings are superior to serum, tonsillar biopsies, conjunctival swabs, and other types of samples in identifying persistently infected pigs ante mortem.9,10 Testing tonsilar crypt exudate by reverse transcription-polymerase chain reaction (RT-PCR) appears to be the most accurate method of identifying persistently infected pigs ante mortem. This method commonly detects PRRSV in more samples and for a longer post-infection period than virus isolation.10,11 A previous study8 has demonstrated the presence of PRRSV using RT-PCR techniques on tonsil or specific lymph nodes at specific times post inoculation during post mortem procedures. We believe that an antemortem tonsil evaluation technique is required to properly evaluate PRRSV persistence.
In a University of Missouri study,8 serological testing by ELISA failed to identify individual sows persistently infected with PRRSV in a predominately seropositive herd that was suffering production losses in the nursery phase of production. Oropharyngeal scrapings (ie, tonsilar crypt exudates) from 54 of 191 clinically normal sows (28.3%) tested positive for PRRSV by RT-PCR. However, PRRSV was isolated from the serum of fewer than half of the 191 sows, suggesting that the prevalence of acute infection was very low. Of the 54 sows positive by RT-PCR, 17 were seronegative by HerdChek PRRS ELISA (Idexx Laboratories, Westbrook, Maine), ie, sample-to-positive (S:P) ratios ≤ 0.4. Nine of the 54 positive sows were seronegative both on the day when samples were positive by RT-PCR and 28 days later, and four of them had S:P ratios < 0.1 on both occasions. These results show that in sows persistently infected with PRRSV, circulating antibodies may not be detectable by ELISA testing even when PRRSV is detectable in tonsilar exudate by RT-PCR.
Using a group of originally PRRSV-naive pigs, either inoculated with a farm-specific strain of PRRSV or vaccinated with a modified live virus (MLV) commercial vaccine, we collected oropharyngeal scrapings over a 5-month period to determine how long PRRSV persists in tonsilar crypt exudates and whether a 90% tonsilar clearance rate is adequate to prevent transmission of PRRSV to naive animals.
Animals, housing, and management
Two isolated pork production sites 64 km apart, each > 8 km from other pigs, were used for this study. On each site, naturally ventilated, monoslope buildings with adjoining concrete aprons were used as grow-finish facilities. All pens were 4.8 × 18 m with solid concrete floors and an open-front loafing area (2.4 × 4.8 m) at one end. Nipple waterers and Osborne round feeders (Osborne Industries Inc, Osborne, Kansas) were provided. Before the study began, both facilities had been depopulated for a minimum of 12 months and had been thoroughly sanitized with a high-pressure power washer (cold water) and disinfected with Virkon S (Dupont, Stone Mountain, Georgia).
Forty-four 3-week-old pigs obtained from a PRRSV-negative source were placed in a single pen in each facility (principal pigs). Each pen was modified for these nursery-age pigs, ie, lined with plywood to prevent drafts, bedded with deep straw, and provided with a heat lamp. At 9 weeks of age, pigs in each building were moved into two clean unmodified pens with 22 pigs per pen and nose-to-nose contact permitted between pens.
For the first 3 weeks, principal pigs were hand-fed daily on mats or on the floor. The segregated-early-weaning diet provided for the first 5 days was replaced by a phase I nursery diet for 7 days, followed by a phase II nursery diet for 14 days, a phase III diet for 21 days, an 18% protein grow-finish I diet for 30 days, and a 16% protein grow-finish II diet for 80 days. Beginning with the phase II nursery diet, Chore-Time bin feeders (CTB Inc, Anderson, Missouri) were provided, each with eight covered feeding spaces.
Daily care was provided by separate management teams so that there was no exchange of personnel between the two sites. Stringent biosecurity practices (including showering and clothing changes between sites) were enforced to prevent viral transmission between groups during sample collection. All equipment was sanitized and separated into two independent sampling kits.
This project was approved by the Office of Animal Resources at the University of Missouri.
Diagnostic testing and inoculation of principal pigs
To confirm PRRSV-negative status on arrival of the principal pigs at the facility, blood samples obtained from all animals were tested by HerdChek PRRS ELISA and by RT-PCR for PRRSV. Blood samples and tonsilar scrapings (obtained as previously described8) were collected from all pigs at approximately 7 weeks of age (Day 0) and tested immediately.
Eighty-eight pigs from a single-source site were transported via a livestock trailer to the study sites. Forty-four pigs were unloaded at the first site and the remaining 44 pigs were then transported to the other site. The pigs were then either inoculated with live PRRSV administered by IM injection (44 pigs, LVI; Site One) or were vaccinated with a modified live virus (MLV) commercial PRRS vaccine (44 pigs, MLV; Site Two). The LVI group were inoculated with 2 mL of serum containing approximately 104 viral particles of University of Missouri Veterinary Medical Diagnostic Laboratory isolate #25544, a farm-specific strain currently used in a serum inoculation program. The MLV groups were vaccinated with Inglevac (Boehringer-Ingelheim, St Joseph, Missouri) according to the directions of the manufacturer.
Blood samples and tonsilar scrapings were again collected on Day 14, and tonsilar scrapings were collected on Days 28, 42, 56, 70, 100, 130, and 160. Blood and tonsilar crypt exudate samples were tested by PCR soon after collection and aliquots were saved for use in a swine bioassay. During the collection period, four pigs died at each site, for reasons unassociated with PRRS. After the 5-month observation period, the buildings were washed and disinfected.
Sentinel pigs
When approximately 90% of the principal pigs at each site tested negative by RT-PCR on tonsilar crypt exudate, six additional PRRSV-naive pigs (sentinels), from the same source as the principal pigs and of approximately the same age and weight, were delivered to each site. Two empty pens in each facility were prepared by thorough washing and disinfection, with an empty pen left as a buffer between the originally occupied pens and the clean pens. Three sentinels were placed in each clean pen, then one pen of principals was moved in with them (23 pigs per pen). Normal socialization behavior assured that the sentinels came into direct contact with oral secretions from every principal pig.
Blood and tonsilar scrapings were collected from all sentinels on arrival, and blood samples were collected 30 days later. Blood samples were tested by ELISA and tonsilar crypt exudate by RT-PCR as described.
Testing tonsilar crypt exudate for PRRSV by RT-PCR
The Taqman PCR was performed on tonsilar crypt exudate as previously described,12 using IDT primers and probe (IDT, Coralville, Iowa) and the Qiagen QuantiTect Probe RT-PCR master mix (Qiagen, Valencia, California) in a Stratagene MX4000 (La Jolla, California).
Nucleotide sequence assembly and alignments were performed using DNAStar software (DNAStar Inc, Madison, Wisconsin). To compare the vaccinal virus and the PRRSV isolated from the vaccinated and the inoculated groups at the conclusion of the study, phylogenetic analyses of nucleotide and amino acid alignments were performed using distance matrix methods (PRODIST, followed by NEIGHBOR) of the DNADIST module within the PHYLIP software package (Phylogeny Inference Package, version 3.5c; University of Washington, Seattle, Washington). Completed tree files were visualized using TreeView 1.5.13
Swine bioassay
Inocula for 20 bioassays were prepared from tonsilar crypt exudate samples collected from the principal pigs on Days 28, 42, 56, 70, 100, and 130. A total of 15 PRRSV-positive pools were created using RT-PCR-positive samples, and five PRRSV-negative pools were created using RT-PCR-negative samples. Each pool was brought up to a total volume of 2.1 mL using phosphate buffered saline, and the resulting homogenates were stored at -80°C. Five additional PRRSV-positive pools were created from RT-PCR-positive serum samples. Bioassay homogenates from MLV and LVI groups were prepared and stored separately.
The 25 PRRS-seronegative bioassay pigs were obtained from the same PRRSV-negative source as the principal and sentinel pigs. Twenty bioassay pigs, housed in a single pen, were each inoculated intramuscularly (IM) in the neck area at 3 weeks of age with 2 mL of a single PRRSV-positive homogenate. Five bioassay pigs injected IM with PRRSV-negative homogenates (2 mL per pig) were housed in an isolated facility (10 km from the nearest pigs) in a pen with a solid concrete floor, deep straw bedding, and a heat lamp. Blood samples collected from all 25 pigs 30 days post inoculation were tested by HerdChek ELISA.
Statistical analysis
The Mixed procedure of SAS version 9.1 (SAS Institute Inc, Cary, North Carolina), using compound symmetry as a variance covariance matrix, was used to analyze PCR data from both the MLV and LVI groups. Survival analysis statistical methods tied to an event (inoculation) were utilized to report this data, creating two right-hand truncated curves that can be analyzed for compound symmetry. The fixed effects of infection method were tested for significance at a level of P < .05 for differences between MLV and LVI groups in the amount of PRRSV RNA detected by quantitative RT-PCR.
Results
Principal pigs
All LVI and MLV pigs were seronegative by ELISA at the beginning of the study, ie, sample:positive (S:P) ratios were < 0.4. All principal pigs became seropositive (ie, S:P ratios ≥ 0.4) beginning with the Day 14 samples and remained seropositive at Day 160.
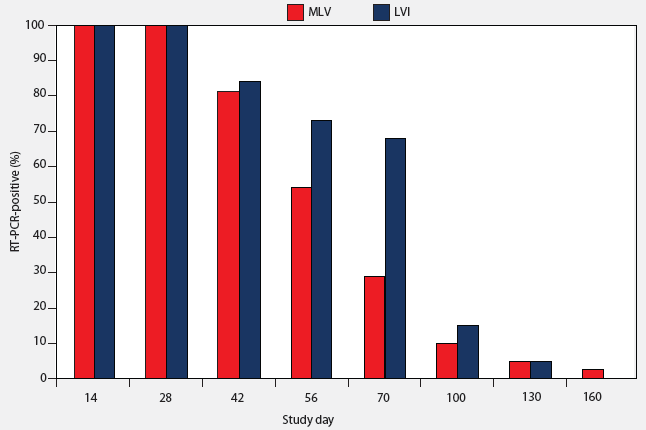
Tonsilar crypt exudate samples from all LVI and MLV groups were negative by RT-PCR on Day 0. On Days 14 to 70, PRRSV was detectable by RT-PCR in tonsilar crypt exudate samples from both inoculated and vaccinated pigs (Table 1), but 90% of the principal pigs had become PCR-negative by Day 130 (Figure 1). A single pig in the MLV group remained PCR-positive at Day 160.
Table 1: Results of reverse transcription-polymerase chain reaction (RT-PCR) testing of tonsilar exudate swabs from pigs either inoculated with live porcine reproductive and respiratory syndrome (PRRS) virus (LVI; 44 pigs) or vaccinated with a modified live virus (MLV) PRRS vaccine (MLV; 44 pigs) at 7 weeks of age (Day 0)*
* All pigs were RT-PCR-negative on tonsilar exudate swabs collected Day 0 before inoculation or vaccination. |
||||||||||||||||||||||||||||||||||||||||||||||||||||
Figure 1: Percent of pigs testing positive by reverse transcription-polymerase chain reaction (RT-PCR) on tonsilar crypt exudate after inoculation on Day 0 at 7 weeks of age with either a live field strain of PRRSV administered in serum by IM injection (LVI; 44 pigs) or vaccinated with modified live virus vaccine according to the directions of the manufacturer (MLV; 44 pigs). Inoculum for the LVI pigs contained approximately 104 viral particles per dose.
|
The amount of PRRSV RNA detected in tonsilar crypt exudate samples was greater for the LVI group on Day 14 (Table 1). This difference was not observed on Day 28 or thereafter.
Sentinel pigs
All 12 sentinel pigs were PRRSV-negative by RT-PCR testing of tonsilar crypt exudate at the time of introduction, and were seronegative by ELISA (ie, S:P ratios < 0.4). both at introduction and 30 days later.
Bioassay pigs
The 25 bioassay pigs were all seronegative when tested by serum ELISA before inoculation with bioassay homogenates. On day 30 post injection, the five pigs injected with PRRSV-negative tonsilar exudate homogenates remained seronegative, but the 15 pigs inoculated with PRRSV-positive tonsilar exudate homogenates and the five pigs inoculated with PRRSV-positive serum pools had seroconverted, ie, ELISA S:P ratios ≥ 0.4.
Similarity of viruses
The vaccinal virus was 100% and 99.5% identical at the nucleotide level with PRRS viruses isolated from the MLV and the LVI groups, respectively.
Discussion
Our first objective was to use an ante mortem test to determine how long the tonsilar crypts remain positive for PRRSV virus after pigs are either inoculated with field virus or vaccinated with a commercial MLV vaccine. The principal pigs in this study were serologically positive until the end of the study (Day 160). However, in 90% of pigs tested, tonsilar crypt exudate was negative by RT-PCR by Day 130, and only one vaccinated pig was still PCR-positive on Day 160, showing that PRRSV (field or vaccine strains) can be harbored in tonsilar crypts for up to 160 days. In agreement with this, Wills et al11 showed that most pigs were PRRSV-negative within 3 to 4 months post inoculation when tested by RT-PCR on tonsilar crypt exudate samples. Wills et al9 also demonstrated that PRRSV was still detected by viral isolation in oropharyngeal scrapings from one of four pigs on day 157 post inoculation.
Our second objective was to determine whether field and vaccine virus differed in their abilities to colonize the tonsil and persist in the tonsilar crypt exudate. The number of PRRSV copies per mL of tonsilar crypt exudate was greater for the LVI group than the MLV group on Day 14, but this difference was not observed at later data collection points. This initial difference may be partially explained by the difference in the inoculum titers, as the LVI inoculum contained 104 viral particles, and this live inoculum may have replicated in the pig following inoculation. However, under the conditions of this study, there was no apparent difference in the abilities of the field and vaccine viruses to colonize the tonsil.
Our final objective was to demonstrate that PRRSV harbored in the tonsilar crypts can be transmitted to naive animals. All sentinel pigs remained seronegative by PRRS ELISA when they were exposed for 30 days to principal pigs when 90% of the principals were PRRSV-negative by RT-PCR on tonsilar crypt exudate samples. However, all 15 bioassay pigs inoculated either with RT-PCR-positive tonsilar crypt exudate or serum obtained from the principal pigs on Days 28 through 130 seroconverted by 30 days post inoculation, substantiating our belief that the viral RNA isolated from the tonsilar crypt exudate represents infectious PRRSV.
Our results differ from those of a previous study8 in which ante mortem tonsilar crypt exudate samples were RT-PCR-positive in serologically negative sows in a commercial sow herd believed to be circulating PRRSV. In that study, 17 of 54 sows (31.5%) were RT-PCR-positive but serologically negative. Factors that might explain the difference in results include the different PRRSV isolates used in the two studies, and infection acquired by different means in pigs of different ages, ie, inoculated and vaccinated nursery pigs and naturally infected adult sows.
In this study, PRRSV was undetectable by RT-PCR testing of tonsilar crypt exudate collected from 79 of the 80 principal pigs 160 days post inoculation or vaccination. This implies that these pigs pose a minimal risk of transmitting the virus after this time. In addition, after 130 days post vaccination, naive pigs exposed to vaccinated and inoculated pigs remained seronegative, suggesting that 130 days after exposure to MLV vaccine or field virus, there is little risk of transmission to naive pigs. However, naive pigs seroconverted after being injected with bioassay samples obtained at 130 days from both the vaccinated and inoculated principal pigs.
The results of this study may have important implications for producers, but will need to be further investigated, as the current design did not allow for multiple replications. The time necessary to eliminate the risk of transmission of PRRSV is at least 130 days post infection, but risk of transmission may still exist in a population of pigs for at least 160 days. Therefore, to eliminate the risk of PRRSV transmission and its possible disease ramifications, animals exposed to PRRSV (or vaccinated with MLV) should be isolated for at least 160 days post exposure. Likewise, after an outbreak of PRRS, a herd should be closed for at least 160 days after the last animal is actively exposed to PRRSV before naive animals (ie, replacement gilts) can be introduced without fear of viral transmission. Results of RT-PCR testing tonsilar crypt exudate samples might be used to assess and minimize the necessary period of isolation or herd closure.
In future studies intended to further simulate a field model associated with circulating PRRSV, a second inoculation at or about 56 days after the first exposure will be used to determine whether re-exposure and subsequent re-colonization of the tonsilar crypts is associated with continued ELISA-positive status.
This ante mortem technique for obtaining tonsilar crypt exudate can be used effectively to quantify PRRSV harbored on the tonsils of infected pigs, and may have additional application in future studies that will use intervening strategies to determine whether the persistent nature of the PRRSV in tonsilar tissue can be altered (eg, by use of feed additives or timed stressful events).
Implications
- Under the conditions of this study, inoculated and vaccinated pigs may remain seropositive by PRRS ELISA beyond the time when they are PRRSV-positive by PCR testing of tonsilar crypt exudate.
- PRRS virus may persist in tonsilar crypt exudate for 160 days post exposure.
- Under the conditions of this study, PRRSV-naive sentinels may remain seronegative 30 days after mixing with a group of inoculated or vaccinated pigs if exposure does not occur until 90% of the infected pigs are PRRSV-negative by PCR on tonsilar crypt exudate samples.
- The rate of PRRSV elimination from the tonsil is the same whether pigs are inoculated with field virus or vaccinated with a commercial MLV vaccine.
- The ante mortem technique for obtaining tonsilar crypt exudate can be used to quantify PRRSV harbored on the tonsils of infected pigs.
Acknowledgements
The authors would like to sincerely thank the National Pork Board in Des Moines, Iowa, for their financial support of this project and Ms Denise Meyer for her analytical assistance. We would also like to thank the producers who took part in the study for the use of their facilities and for providing daily care for these pigs.
References
1. Plagemann PGW. Lactate dehydrogenase-elevating virus and related viruses. In: Fields B, Knipe DM, Howley PM, eds. Fields Virology. 3rd ed. Philadelphia, Pennsylvania: Lippincott-Raven; 1996:1105–1120.
2. Albina E, Madec F, Cariolet R, Torrison J. Immune response and persistence of the porcine reproductive and respiratory syndrome virus in infected pigs and farm units. Vet Rec. 1994;134:567–573.
3. Allende R, Laegreid WW, Kutish GF, Galeota JA, Wills RW, Osorio FA. Porcine reproductive and respiratory syndrome virus: Description of persistence in individual pigs upon experimental infection. J Virol. 2000;74:10834–10837.
*4. Benfield DA, Nelson J, Rossow KD, Rowland RR, Lawson SR, Steffen M, Collins M. Pathogenesis and persistence of PRRS. Proc AD Leman Swine Conf. 1998;169–171.
5. Christopher-Hennings J, Nelson EA, Hines RJ, Nelson JK, Swenson SL, Zimmerman JJ, Chase CCL, Yaeger MJ, Benfield DA. Persistence of porcine reproductive and respiratory syndrome virus in serum and semen of adult boars. J Vet Diagn Invest. 1995;7:456–464.
6. Sur JH, Cooper VL, Galeota JA, Hesse RA, Doster AR, Osorio FA. In vivo detection of porcine reproductive and respiratory syndrome virus RNA by in situ hybridization at different times postinfection. J Clin Microbiol. 1996;34:2280–2286.
7. Yoon IJ, Joo HS, Christianson WE, Morrison RB, Dial GD. Persistent and contact infection in nursery pigs experimentally infected with porcine reproductive and respiratory syndrome (PRRS) virus. Swine Health Prod. 1993;1(4):5–8.
8. Kleiboeker SB, Lehman J, Fangman TJ. Concurrent use of reverse transcription-polymerase chain reaction testing of oropharyngeal scrapings and paired serological testing for detection of porcine reproductive and respiratory syndrome virus infection in sows. J Swine Health Prod. 2002;10:251–258.
9. Wills RW, Zimmerman JJ, Yoon KJ, Swenson SL, McGinely MJ, Hill HT, Platt KB, Christopher-Hennings J, Nelson EA. PRRS virus: a persistent infection. Vet Microbiol. 1997;55:231–240.
10. Horter D, Pogranichniy R, Chang CC, Evans RB, Yoon KJ, Zimmerman JJ. Characterization of the carrier state in porcine reproductive and respiratory syndrome virus infection. Vet Microbiol. 2002;86:213–228.
11. Wills RW, Doster AR, Galeota JA, Sur JH, Osorio FA. Duration of infection and proportion of pigs persistently infected with porcine reproductive and respiratory syndrome virus. J Clin Microbiol. 2003;41:58–62.
12. Kleiboeker SB, Schommer SK, Lee SM, Chittick W, Polson D. Simultaneous detection of North American and European porcine reproductive and respiratory syndrome virus using real-time reverse transcriptase-PCR. J Vet Diagn Invest. 2005;17:165–170.
13. Page RDM. TREEVIEW: an application to display phylogenetic trees on personal computers. Comp Appl Biosci. 1996;12:357–358.
*Non-refereed reference.