| Case report | Peer reviewed |
SummaryAn outbreak of reproductive failure associated with porcine circovirus type 2 (PCV2) occurred in a closed, PCV2-naive, specific-pathogen-free herd in Iowa in 2009. Elimination of infectious PCV2 from the breeding-herd site, the outcome after repopulation, and the attempt to derive PCV2-negative animals by offsite segregation are summarized. Clinical signs were limited to an increased incidence of mummified fetuses. After confirmation of PCV2-associated lesions in the fetuses and PCV2 viremia in dams, the herd was depopulated. Cleaning and disinfection of the premises prior to repopulation included removal of gross organic material, exposure of equipment to natural UV light, multiple applications of disinfectant, and application of paint or sealer to porous surfaces. During the 63-day clean-up period, no pigs were on the site. An improved biosecurity plan was implemented. The herd was repopulated and a PCV2-naive population had remained PCV2-negative for 20 months at the time of writing. Attempts to derive PCV2-negative pigs from the positive herd following offsite segregation were unsuccessful. The combination of a multistep cleaning and disinfection protocol with a strict biosecurity plan can result in the maintenance of PCV2-naive animals on a previously contaminated site. | ResumenUn brote de falla reproductiva asociada con circovirus porcino tipo 2 (PCV2) ocurrió en una piara cerrada y previamente libre de PCV2 y patógenos específicos en Iowa en el 2009. En este artículo, se resumen la eliminación del PCV2 infeccioso del pie de cría, el resultado de la repoblación, y el intento de producir animales negativos al PCV2 mediante la segregación fuera de sitio. Los signos clínicas se limitaron a un incremento en la incidencia de fetos momificados. Después de la confirmación de las lesiones asociadas con el PCV2 en los fetos y viremia de PCV2 en las hembras se despobló el hato. La limpieza y desinfección de las instalaciones antes de la repoblación incluyeron la eliminación de material orgánico, la exposición de equipo a luz UV natural, aplicaciones múltiples de desinfectante, y la aplicación de pintura ó sellador a las superficies porosas. Durante el periodo de limpieza de 63 días, no hubo cerdos en el sitio. Se implementó un plan de bioseguridad mejorado. El hato se repobló y la población ha permanecido libre de PCV2 durante 20 meses y hasta el momento de la preparación de este manuscrito. Los intentos de producción de cerdos negativos al PCV2 provenientes del hato positivo después de la segregación fuera del sitio fueron infructuosos. La combinación del protocolo de desinfección y una limpieza de pasos múltiples con un estricto plan de bioseguridad pueden resultar en el mantenimiento de animales libres de PCV2 en un sitio previamente contaminado. | ResuméUne épidémie de problèmes reproducteurs associés au circovirus porcin de type 2 (PCV2) est survenue en Iowa en 2009 dans un troupeau fermé, exempt d’agent pathogène spécifique et naïf pour le PCV2. L’élimination de PCV2 infectieux du site, le résultat après la repopulation, et la tentative d’obtenir des animaux PCV2 négatifs par ségrégation hors-site sont résumés. Les signes cliniques étaient limités à une augmentation de l’incidence de fœtus momifiés. Après confirmation de lésions associées au PCV2 dans les fœtus et de virémie à PCV2 chez les mères, une dépopulation du troupeau a été faite. Le nettoyage et la désinfection des lieux avant une repopulation incluaient l’enlèvement du matériel organique visible, l’exposition de l’équipement aux rayons UV naturels, des applications multiples de désinfectant, et l’application de peinture ou de scellant sur les surfaces poreuses. Aucun porc n’était présent sur les lieux pendant les 63 jours de la période de nettoyage. Un plan de biosécurité amélioré a été mis en place. La repopulation du troupeau a été effectuée et la population naïve pour PCV2 est demeurée négative pour PCV2 jusqu’au moment de la rédaction du présent rapport, soit 20 mois plus tard. Des tentatives d’obtenir des animaux négatifs pour PCV2 à partir du troupeau positif par ségrégation hors-site se sont avérées infructueuses. La combinaison d’étapes multiples de nettoyage et de désinfection avec un plan strict de biosécurité peut se solder par le maintient d’animaux naïfs pour PCV2 sur un site préalablement contaminé. |
Keywords: swine, porcine circovirus type 2, disinfection, biosecurity, PCV2, case report
Search the AASV web site
for pages with similar keywords.
Received: June 4, 2010
Accepted: November 19, 2010
Porcine circovirus type 2 (PCV2) is a small, non-enveloped, single-stranded DNA virus which emerged in the 1990s as an economically important swine pathogen.1 The first report of PCV2 reproductive disease occurred in a commercial, 450-head herd composed entirely of first-parity dams.2 The timing of infection was not determined in that case, and clinical signs on the farm included late-term abortions (losses after 13 weeks of gestation), decreased farrowing rates, and increased stillborns and mummified fetuses; no clinical signs were reported in dams. In one of nine examined fetuses, myocarditis and abundant PCV2 antigen were identified in fetal myocardium. Other common pathogens associated with reproductive disease were not detected.2 Multiple case reports on PCV2-associated reproductive failure have subsequently been described in seropositive herds3,4 or in start-up herds or gilt populations that may have been serologically naive to PCV2.5-8 Common clinical features included increased numbers of mummified fetuses and stillborns at parturition, no or low numbers of dams exhibiting clinical signs, and resolution of clinical signs between 2 and 5 months following initial detection.
Porcine circovirus type 2 is quite stable in vitro,9,10 and many disinfectants do not completely eliminate the virus under in vitro conditions.11-13 To our knowledge, no documented information exists on facility disinfection procedures for establishment and maintenance of a PCV2-negative herd. However, a procedure for room decontamination used successfully by researchers at Iowa State University to disinfect rooms between PCV2 animal-inoculation studies was recently described.14
A natural outbreak of PCV2-associated disease occurred in a high-health population previously known to be free of PCV2, and the procedure used to disinfect the premises, the outcome after repopulation, and an attempt to derive PCV2-negative animals by offsite segregation are described.
Building descriptions
The source herd was housed in a facility located in Iowa, originally built in the 1970s. The facility was composed of five 22.8 m × 15.2 m buildings in close proximity to each other (15 to 30 m) with an associated waste lagoon. An uncovered concrete walkway connecting the individual buildings had been installed after herd depopulation following the 2009 outbreak. A diagram of the facility after May 2009 is shown in Figure 1.
Figure 1: Diagram of a swine research breeding facility following complete depopulation in May 2009 after an outbreak of reproductive failure associated with porcine circovirus type 2. 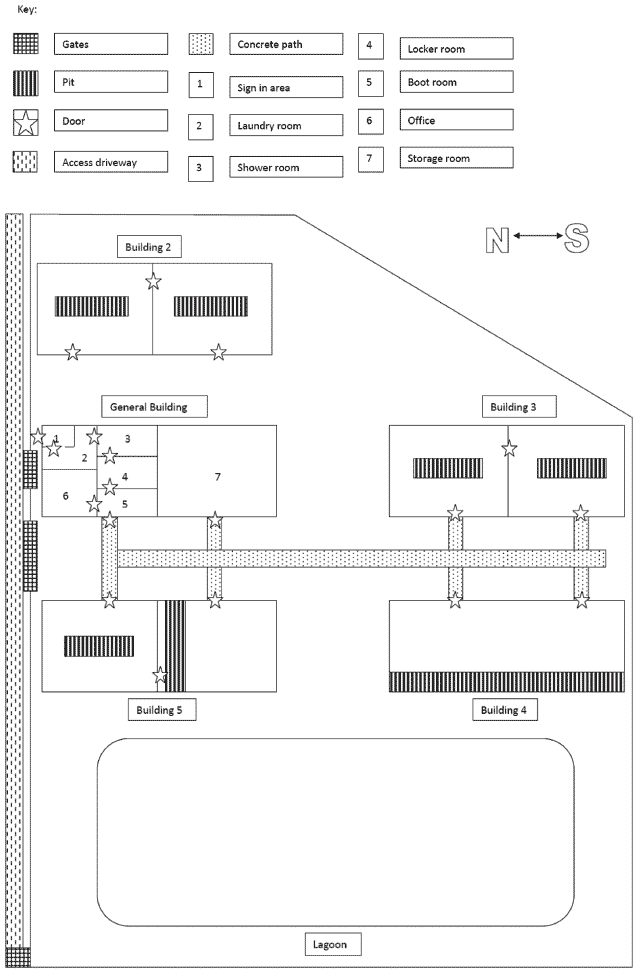 |
General services building. Building 1, used as the general services building, was divided into two sides by a concrete wall. One side was used as the entrance to the facility with office space, shower, laundry, and locker room, and the second side provided storage for feed and equipment and tools for building maintenance. Building 1 had one door for entry into the sign-in area, a door that led out from the office area, and a garage door for entry into a feed and equipment storage area.
Animal buildings. Buildings designated 2 through 5 were used for animal housing. The animal buildings were completely enclosed, power ventilated, and had partially slatted floors with pull-plug drains. Buildings 2, 3, and 5 each had a center concrete wall which divided the building into halves lengthwise; each half had a separate access point and a separate manure pit (Figure 1). The manure pits were 1.2 m wide, 3.4 m long, and 0.9 m deep. The remaining floor space had a sloped solid floor. Building 4 had one manure pit 1.2 m wide, 22.8 m long, and 0.9 m deep.
Herd description and housing prior to depopulation in May 2009
Naive sows and gilts were placed on site approximately 2 years prior to the described outbreak. All animals came from a PCV2-naive herd, as determined by PCV2 serological and polymerase chain reaction (PCR) testing, and all were immediately tested by PCV2 serology and PCR after arrival at the facility. No new animals entered the facility from the time of initial population in December 2007 until depopulation in May of 2009. Sows were cross-bred, specific-pathogen-free animals naive for PCV2, porcine reproductive and respiratory syndrome virus (PRRSV), swine influenza virus (SIV), and porcine parvovirus (PPV), as determined by periodically performed serological testing (PCV2, PRRSV, SIV, and PPV) and PCR testing (PCV2, PRRSV) on sows of mixed parities and their offspring. Sows and gilts were housed in Buildings 2 and 3 during gestation and moved into Building 5 prior to farrowing. Building 3 contained mature boars as well as gestating sows. Building 4 was primarily used as a nursery unit. During the observation period, a total of 38 breeding animals were onsite (12 in Building 2, 13 in Building 3, two in Building 4, and 11 in Building 5).
PCV2-associated reproductive disease outbreak
Outbreak detection and confirmation
Details on the timeline of the PCV2 outbreak are summarized in Figure 2. The outbreak was first noted on January 29, 2009, when a portion of weaned 2-week-old pigs from Building 4 were routinely screened and were positive for anti-IgG PCV2 antibodies when tested using a previously described ELISA.15 Subsequently, the presence of anti-PCV2 antibodies in the samples was confirmed by an in-house immunofluorescent antibody (IFA) assay,16 and PCV2 DNA was detected by using a quantitative real-time PCR with a detection limit of 1 × 103 copies per mL.17 Serum samples were collected from all sows, and a mixture of seropositive nonviremic animals, seropositive viremic animals, and seronegative viremic animals were detected in Buildings 2, 3, and 5. Neither anti-PCV2 antibodies nor PCV2 DNA were detected in the serum of the two gilts housed by themselves in one half of Building 4. The exact timing of infection was not deduced from this data. However, there were higher proportions of sows in which PCV2 DNA was detected without detectable anti-PCV2 antibody in Buildings 2 (four of 12; 33.3%) and 3 (four of 13; 30.8%) than in Building 5 (one of 11; 9.1%). Infection was apparently most recent in Buildings 2 and 3 and of longer duration in Building 5.
Figure 2: Timeline of events after detection of an outbreak of reproductive failure associated with porcine circovirus type 2 (PCV2) in a swine research breeding facility and repopulation of the facility with a PCV2-naive research breeding herd. 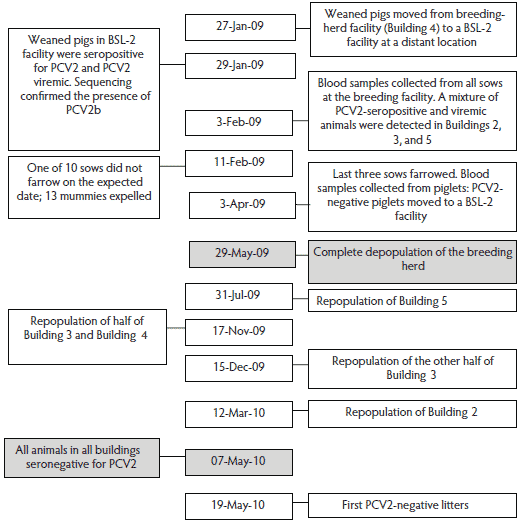 |
To further characterize the PCV2 strain that infected the herd, PCV2 open reading frame 2 (ORF2) sequencing was performed on extracted DNA from two sow serum samples and two piglet serum samples using a nested PCR as previously described.18 The PCR products were purified using the QIAquick PCR purification kit (Qiagen, Valencia, California) per manufacturers’ instructions and sequenced at the Iowa State University DNA facility. Sequences were analyzed with Sequence Scanner software version 1.0 (Applied Biosystems, Foster City, California) and compared with four common PCV2a and PCV2b strains using the basic local alignment search tool (BLAST),19 and the isolate was identified as PCV2b (100% homology with GenBank Accession No. EU340258).
Reproductive parameters and clinical observations in breeding animals from May 2008 to April 2009
Farrowing data from the 38 sows between May 2008 and April 2009 were recorded by Iowa State University Laboratory Animal Resources personnel. Reproductive failure was evident in one sow that farrowed a term litter composed of 13 mummies in February 2009. Microscopic evaluation of the mummified fetal tissues revealed multifocal, severe myocardial necrosis with mineralization. Abundant PCV2 antigen was detected by immunohistochemistry (IHC) in the fetal myocardium.20 One sow was euthanized in May 2008 due to hind-limb paresis associated with a vertebral abscess. No other clinical signs were reported in the breeding herd during this time period. The approximate time of PCV2 infection of the breeding herd was between December 2008 and January 2009. The PCV2 status of 10 sows at midgestation (approximately 57 days; February 3, 2009) and their litter characteristics at farrowing (number of mummified fetuses, stillborns, and born-alive piglets) are summarized in Table 1. Although nine of the 10 sows were exposed to PCV2 as determined by PCR and serology data, reproductive performance appeared normal.
Table 1: Dam PCV2 antibody and viremia status at approximately 57 days of gestation (February 3, 2009) and characteristics of their litters at the time of farrowing in a swine research breeding herd naturally infected with PCV2
* ELISA previously described.15 Sample-to-positive ratios presented. † IFA previously described.16 ‡ Determined by quantitative real-time polymerase chain reaction. PCV2 = porcine circovirus type 2; ELISA = enzyme-linked immunosorbent assay; IFA = immunofluorescent assay. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Removal of PCV2 from the farm environment
Facility biosecurity protocols
Access to the facility was through a gated entrance that was locked when employees were not present. An additional perimeter fence enclosed the swine facility, where a biosecurity sign was posted. To further limit the number of personnel that had access, all animal buildings were locked at all times. At entry into the facility, a change of clothes was required. The only vehicles granted access to the swine facility were a snow blower and a lawn mower, both nonspecific to the swine facility, which were used for property maintenance. All equipment needed in the facility was disinfected with Virkon S (Dupont, Pharmacal Research Laboratories, Inc, Naugatuck, Connecticut) prior to entrance through the biosecurity gate. Upon access to the facility, the use of foot baths was required between all animal buildings. A custom-fitted system for air filtration was installed on the outside of air inlets in each building using air filters (3M Filtrete Micro Particle Reduction Filter 700; 3M Co, Ames, Iowa) and custom frames. Pits were emptied once per week into a nearby lagoon. The lagoon was emptied once per year in the fall by facility employees and contents were spread onto an adjacent field. For rodent control, four bait boxes per building were filled with a brodifacoum product (Havoc Rodenticide Bait Pack; Hacco, Inc, Randolph, Wisconsin). For bird control, foam insulation (Great Stuff insulating foam; Dow Chemical Company, Midland, Michigan) was placed into openings between the roof and walls. Specific differences in the biosecurity protocol concerning animal movements between buildings, personnel movements between buildings, semen usage, feed delivery, equipment disinfection, visitor entry procedures, air management, manure-pit management, and daily chores prior to the PCV2 outbreak and following repopulation of the herd are summarized in Table 2.
Table 2: Outline of specific biosecurity protocols used at a research swine breeding facility site prior to an outbreak of reproductive failure associated with PCV2 infection and after repopulation of the facility with PCV2-naive animals
* Tractor also used in cattle, horse, and small-ruminant facilities in close proximity to the swine facility. PCV2 = porcine circovirus type 2; PCR = polymerase chain reaction. |
Disinfection of the premises
General services building. All disposable supplies were discarded, including shelving units, cleaning supplies, clothing and boots, and all movable equipment (ie, facility washer and dryer) was moved outside the building. This was followed by thorough cleaning and disinfection of the building and equipment. Specifically, floors were scrubbed with a degreaser (PRL-Grease Free; Pharmacal Research Laboratory Inc, Naugatuck, Connecticut) followed by rinsing and disinfection with a chloride compound (Clorox Bleach; The Clorox Company, Oakland, California). The surface of the washer and dryer were cleaned with general household cleaning products and exposed to natural UV sunlight. In addition, a rinse cycle of hot water that included bleach was run through the washing machine prior to its replacement in the cleaned facility. New shelving units for clothing, cleaning supplies, and other supplies were placed in the office. All tools were cleaned with water and sprayed with disinfectant (Virkon S).
Buildings 2 through 5. Animals were removed from Buildings 2 through 5 starting on May 14, 2009. Complete depopulation of the site was achieved on May 29, 2009. Steps used in the cleaning and disinfection of the animal facilities are outlined in Table 3.
Table 3: Stepwise cleaning and disinfection protocol used in Buildings 2 to 5 of a research swine breeding facility following an outbreak of reproductive failure associated with porcine circovirus type 2 infection
|
Monitoring of the disinfection success of the premises
Following application of disinfectant in the cleaning and disinfection protocol (Table 3, Step 4), 10 swabs of each building were collected from areas likely to contain virus (eg, pit, floor, louvers). The surface swabs (polyester-tipped swab; Fisher Scientific Inc, Pittsburgh, Pennsylvania) were collected, each placed into 1 mL of sterile saline (0.9% sodium chloride solution; Fisher Scientific Inc) and stored at -80°C until tested by quantitative real-time PCR for presence and amount of PCV2 DNA. Porcine circovirus type 2 DNA was identified on a plastic sort panel, in two of 10 samples taken from the office (a shelf and under a computer cabinet), the door and floor of Building 5, the drain and floor of Building 4, the floor and drain of Building 2, and in several swabs from the pit grate and pit floor in Buildings 2 through 5. The mean log10 genomic PCV2 DNA per mL ± standard error for these positive samples was 4.48 ± 0.14. Other locations sampled where PCV2 DNA was not detected included gating, light switches, foot baths, water nipples, pig transport carriers, the desk in the office area, the storage room floor, louvers, electrical boxes, and fan inlets.
Repopulation of the site
A single building (Building 5) (Figure 1) was initially repopulated with PCV2-naive animals (negative for anti-PCV2 IgG antibodies15 and PCV2 DNA in serum) at the end of July 2009 (63 days after depopulation). Blood samples were collected monthly and tested for evidence of seroconversion to PCV2 by ELISA.15 Due to an increase in size of the animals and subsequently a need for more space, some animals were placed in Buildings 3 and 4 in November of 2009. Animals were placed in Building 2 on March 12, 2010. As of September 2010, the populations of the buildings were as follows: 21 open gilts, three pregnant sows, and one boar in Building 2; six lactating sows, two pregnant gilts, five open sows, and one boar in Building 3; five open sows in Building 4; and eight open sows and one boar in Building 5. Neither viremia nor seroconversion to PCV2 were detected in any of the 53 animals up to the time of writing, approximately 20 months since repopulation of the herd. Testing has been performed on a rotating basis, with every animal tested at least once every 2 months. The first PCV2-naive litters were born in May 2010 (Figure 2).
Attempt to derive negative animals from PCV2-infected sows during the outbreak
Piglets from sows that farrowed at the beginning of April 2009 were blood sampled 13 days post farrow (DPF). Of these, 15 female piglets from four litters that were not PCV2-viremic at the time of sampling were weaned at 22 to 27 days of age and placed into a Biosafety Level 2 (BSL-2) facility at a different site. The pigs were housed in four separate rooms by litter. Each room had a solid concrete floor, a separate ventilation system, and one nipple drinker. The pigs were fed a balanced, pelleted, complete-feed ration free of antibiotics and animal proteins other than whey (Nature’s Made; Heartland Co-op, Cambridge, Iowa) once a day. Piglets were sampled monthly; serum was used to determine levels of anti-PCV2 IgG and the PCV2 viremia status of the animals by ELISA15 and PCR,17 respectively. This experimental protocol was approved by the Iowa State University Institutional Animal Care and Use Committee. Anti-PCV2 antibodies were detectable in all piglets from all litters at DPF 13. As a numeric trend toward decline of antibodies was noted between DPF 13 and DPF 204, these antibodies were assumed to be of maternal origin (Figure 3). At DPF 145, eight pigs were serologically negative, and anti-PCV2 antibodies declined to a sample-to-positive (S:P) ratio < 0.2 in the remaining two animals by DPF 176 (Figure 3). Porcine circovirus type 2 DNA was not detected in the piglets at 13 DPF; however, by 21 DPF, in one pig in each of three rooms, samples contained detectable amounts of PCV2 DNA and those pigs were subsequently removed. Any additional pig that was found to be positive for PCV2 DNA by PCR throughout the duration of the study was immediately removed. By DPF 204, 100% of the remaining piglets were PCR-positive for PCV2 DNA (Figure 3). Therefore, the study was terminated and the pigs were euthanized. The entire PCV2 ORF2 from one piglet in each room was sequenced as described and was 100% identical to the PCV2 ORF2 recovered from the sows during the outbreak, thus confirming that the piglets were likely infected at the time of offsite transfer and segregation in the BSL-2 facility.
Figure 3: In the swine research breeding facility described in Figures 1 and 2, piglets from PCV2-infected sows that farrowed at the beginning of April 2009 were blood sampled 13 days post farrow. Fifteen female piglets from four litters not PCV2-viremic at the time of sampling were weaned at 22 to 27 days of age and placed in a Biosafety Level 2 facility at a different site and housed in four different rooms by litter. Monthly serum samples were tested for anti-PCV2 IgG and PCV2 viremia by ELISA15 and quantitative real-time polymerase chain reaction (PCR), respectively. The graph shows mean pig anti-PCV2 sample-to-positive (S:P) ratio (line) and percentage of PCV2 PCR-positive piglets (bar) at different days post farrowing. Mean values were generated by the following numbers of pigs tested at each time point: 15 pigs at days 13, 21, and 51; 13 pigs at day 84; 12 pigs at day 114; 10 pigs at day 145; six pigs at day 176; and six pigs at day 204. 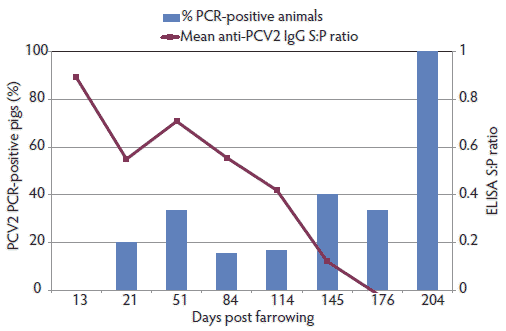 |
Discussion
A natural outbreak of PCV2-associated reproductive disease occurred in a closed high-health population, and the procedure used to disinfect the premises before repopulation with PCV2-naive animals, the outcome after repopulation, and the attempts to derive PCV2-negative animals by offsite segregation were summarized. The PCV2 outbreak in the research breeding herd was originally detected on the basis of serological evaluation of recently weaned piglets and later confirmed by IHC staining for PCV2 in mummified fetuses, detection of anti-PCV2 antibodies by IFA and ELISA, and detection of PCV2 DNA by PCR on serum samples from sows. Viremia was detected in eight mid-gestation sows (approximately 57 days), and PCV2 antigen was demonstrated by IHC on mummies from one sow. This is consistent with previous studies involving experimental PCV2 infection of sows indicating that early infection (1 to 35 days of gestation) resulted in embryonic death,21 irregular returns to estrus,22 pseudopregnancy,23 or small litter sizes;2 infection at mid-gestation (35 to 70 days) resulted in mummified fetuses and abortion;21 and infection during late gestation (70 to 115 days) resulted in mummified fetuses,24 stillborns,25 weak-born piglets,4 delayed farrowing,26 normal litters,27 or abortion.28
The exact timing of PCV2 infection in the research breeding herd remains undetermined. It can be assumed that initial infection occurred sometime after December 11, 2008 (last batch of naive animals taken from the herd), and before January 27, 2009 (first detection of PCV2 in weaned pigs). Similarly, it is difficult to retrospectively determine the building into which PCV2 was first introduced. However, using a higher proportion of sows with anti-PCV2 antibodies as the criterion, the introduction of PCV2 to the site is assumed to have occurred in Building 5. The source strain of infection was a PCV2b isolate. Similar isolates (ie, with 100% homology to the PCV2 recovered from the outbreak, determined by ORF2 sequencing) were used commonly in a research laboratory 8 km to the north, and the farm was frequently visited by people working at the research facility. Horizontal transmission likely occurred from either contaminated equipment or people. However, there are a large number of possible routes by which horizontal transmission may have occurred due to the design of the facility and concurrent responsibilities of personnel who visited the research herd.
Attempts to derive PCV2-negative animals from the herd after the outbreak by offsite segregation by litter were unsuccessful. The inability to derive negative pigs was likely due to the presence of an active PCV2 outbreak and persistent PCV2 infection of the piglets, with viral replication beginning as levels of maternal antibodies declined.
Major risk factors that were identified after the outbreak included frequent movement of animals and people between buildings with minimal biosecurity; failure to maintain a shower-in, shower-out facility; preparation of semen extender at an off-site facility; use of a common tractor between the swine facility and other areas of the farm; lack of concrete paths between buildings; and transport of feed directly into the facility. To address these issues, specific changes were made to the facility itself and to the way in which it was operated. The major changes included enforcement of strict bio-security protocols for movement of people and equipment into and on the facility, addition of concrete paths between buildings to reduce organic contamination of boot baths, installation of a building for feed fumigation, and purchase of PCV2-negative semen (determined by PCV2 PCR testing of each semen batch). Another minor change included addition of air filters over outlets. This change was considered minor, as only one site containing pigs (approximately 100 sows) was within an 8.1-km radius of this site. In addition, while little information exists on aerosol transmission of PCV2, in the single study which evaluated the presence of PCV2 DNA in aerosol samples, PCV2 DNA was not detected.29 The risk of PCV2 transmission from rodents or birds was also considered low, and modifications to the current rodent-control protocol were not made. Previous studies have demonstrated PCV2 replication in mice,30 and a 2010 study reported that 65% and 23.8% of the mice and rodents from PCV2-infected swine premises were PCV2-positive, respectively.31 However, PCV2 was not detected in rodents outside of the premises.31 This information, combined with the distance to the nearest swine farm, makes transmission from rodents unlikely. To date, no information exists, to the authors’ knowledge, on whether indirect transmission of PCV2 can occur between avian and porcine species. However, due to the host specificity of circoviruses within avian species,32 transmission by this route seems unlikely.
Disinfection of a facility contaminated with PCV2 is an arduous task. Porcine circovirus type 2 is known to be shed by numerous routes, including nasal and oral secretions, urine, and feces.33 In addition, viremia can persist in animals for extended periods. Porcine circovirus type 2 viremia was previously reported in pigs for 140 days post infection.34 Porcine circovirus type 2 is transmitted both by horizontal35 and vertical routes.36 The virus is also extremely stable,9,10 and many disinfectants do not completely eliminate the virus under in vitro conditions.11-13 Clinical disease associated with PCV2 in breeding herds is rarely observed and typically resolves, after exposure of a potentially naive population to PCV2, between 8 and 20 weeks following the initial detection.2,25,37 Vaccination programs are highly effective in reducing mortality associated with PCV2 infection;38 however, they do not eliminate shedding of PCV2. On the basis of the combination of the above factors, PCV2-naive breeding herds are extremely rare. However, high-health animals free of common viruses and bacteria are required for researchers to further advance understanding of the pathology and epidemiology of swine pathogens. Therefore, documentation of the ability to successfully eliminate PCV2 from a farm is important.
In this study, the combination of thorough removal of organic material using a detergent and exposure of equipment to natural UV light and multiple applications of a disinfectant were used. In addition, all disposable equipment was discarded and surfaces which would likely retain virus were either painted or sealed. Finally, the facility remained without animals for 63 days. As downtime was combined with thorough cleaning and disinfection, it is unknown whether downtime alone would have led to a similar elimination of PCV2 from the premises. However, detection of PCV2 DNA within the pits after thorough cleaning and disinfection suggests that downtime alone would not be enough to prevent transmission from a contaminated building to naive animals. After using the described decontamination protocol and enforcing a strict biosecurity protocol, the repopulated herd has remained PCV2-naive (determined by routine PCR and ELISA screening) for at least 20 months at the time of writing. The depopulation and repopulation procedure described in this manuscript could be implemented by conventional swine farms if a source of known PCV2-free pigs is available.
Implications
• In natural outbreaks of PCVAD in high-health breeding herds, the main clinical sign is likely to be increased numbers of mummified fetuses.
• Factors including frequent movement of animals and people between sites and buildings with minimal biosecurity, lack of maintaining a shower-in-shower-out facility, preparation of semen extension at an off-site facility, use of a common tractor between the swine facility and other areas of the site, and the lack of concrete paths between barns are potential risk factors for PCV2 transmission and should be modified following natural outbreaks of PCV2 in high-health herds.
• After depopulation, implementation of a multistep cleaning and disinfection protocol with a strict biosecurity plan can result in maintenance of PCV2-naive animals on a previously contaminated site.
• Offsite segregation by litter to derive PCV2-negative replacement animals from high-health breeding herds during natural outbreaks of PCV2 is not successful.
Acknowledgements
The authors would like to acknowledge the many Laboratory Animal Resources personnel, especially Roger King, who carried out the cleaning and disinfection of the swine facility, and Paul Thomas and Dr HuiGang Shen for assistance with laboratory and animal work.
References
1. Harding JCS, Clark EG, Strokappe JH, Willson PI, Ellis JA. Postweaning multisystemic wasting syndrome: epidemiology and clinical presentation. Swine Health Prod.1998;6:249–254.
2. West KH, Bystrom JM, Wojnarowicz C, Shantz N, Jacobson M, Allan GM, Haines DM, Clark EG, Krakowka S, McNeilly F, Konoby C, Martin K, Ellis JA. Myocarditis and abortion associated with intrauterine infection of sows with porcine circovirus 2. J Vet Diagn Invest. 1999;11:530–532.
*3. Janke B. Case report: Porcine circovirus as a cause of reproductive problems. Proc Iowa Vet Med Assoc. Ames, Iowa. 2000;101.
*4. Nielsen J, Ladekjær-Hansen AS, Bille-Hansen V, Lohse L, Bøtner A. PCV-associated disease following intrauterine infection. Proc IPVS. Hamburg, Germany. 2004;1:14.
5. Brunborg IM, Jonassen CM, Moldal T, Bratberg B, Lium B, Koenen F, Schönheit J. Association of myocarditis with high viral load of porcine circovirus type 2 in several tissues in cases of fetal death and high mortality in piglets. A case study. J Vet Diagn Invest. 2007;19:368–375.
6. Mauch C, Bilkei G. Porcine circovirus (PCV) associated losses in pregnant gilts. Pig J. 2004;53:69–74.
7. Mikami O, Nakajima H, Kawashima K, Yoshii M, Nakajima Y. Nonsuppurative myocarditis caused by porcine circovirus type 2 in a weak-born piglet. J Vet Med Sci. 2005;67:735–738.
8. Pittman JS. Reproductive failure associated with porcine circovirus type 2 in gilts. J Swine Health Prod. 2008;16:144–148.
9. O’Dea MA, Hughes AP, Davies LJ, Muhling J, Buddle R, Wilcox GE. Thermal stability of porcine circovirus type 2 in cell culture. J Virol Methods. 2008;147:61–66.
10. Welch J, Bienek C, Gomperts E, Simmonds P. Resistance of porcine circovirus and chicken anemia virus to virus inactivation procedures used for blood products. Transfusion. 2006;46:1951–1958.
11. Kim HB, Lyoo KS, Joo HS. Efficacy of different disinfectants in vitro against porcine circovirus type 2. Vet Rec. 2009;164:599–600.
12. Martin H, Le Potier MF, Maris P. Virucidal efficacy of nine commercial disinfectants against porcine circovirus type 2. Vet J. 2008;177:388–393.
13. Royer R, Nawagitgul P, Halbur PG, Paul PS. Susceptibility of porcine circovirus type 2 to commercial and laboratory disinfectants. J Swine Health Prod. 2001;9:281–284.
14. Gillespie J, Opriessnig T, Meng XJ, Pelzer K, Buechner-Maxwell V. Porcine circovirus type 2 and porcine circovirus-associated disease. J Vet Intern Med. 2009;23:1151–1163.
15. Nawagitgul P, Harms PA, Morozov I, Thacker BJ, Sorden SD, Lekcharoensuk C, Paul PS. Modified indirect porcine circovirus (PCV) type 2-based and recombinant capsid protein (ORF2)-based enzyme-linked immunosorbent assays for detection of antibodies to PCV. Clin Diagn Lab Immunol. 2002;9:33–40.
16. Pogranichniy R, Yoon KJ, Harms PA, Swenson SL, Zimmerman JJ, Sorden SD. Characterization of immune response of young pigs to porcine circovirus type 2 infection. Viral Immunol. 2000;13:143–153.
17. Opriessnig T, Yu S, Gallup JM, Evans RB, Fenaux M, Pallares F, Thacker EL, Brockus CW, Ackermann MR, Thomas P, Meng XJ, Halbur PG. Effect of vaccination with selective bacterins on conventional pigs infected with type 2 porcine circovirus. Vet Pathol. 2003;40:521–529.
18. Opriessnig T, McKeown NE, Zhou EM, Meng XJ, Halbur PG. Genetic and experimental comparison of porcine circovirus type 2 (PCV2) isolates from cases with and without PCV2-associated lesions provides evidence for differences in virulence. J Gen Virol. 2006;87:2923–2932.
19. Zhang Z, Schwartz S, Wagner L, Miller W.
A greedy algorithm for aligning DNA sequences.
J Comput Biol. 2000;7:203–214.
20. Sorden SD, Harms PA, Nawagitgul P, Cavanaugh D, Paul PS. Development of a polyclonal-antibody-based immunohistochemical method for the detection of type 2 porcine circovirus in formalin-fixed, paraffin-embedded tissue. J Vet Diagn Invest. 1999;11:528–530.
21. Kim J, Jung K, Chae C. Prevalence of porcine circovirus type 2 in aborted fetuses and stillborn piglets. Vet Rec. 2004;155:489–492.
22. Mateusen B, Maes DG, Van Soom A, Lefebvre D, Nauwynck HJ. Effect of a porcine circovirus type 2 infection on embryos during early pregnancy. Theriogenology. 2007;68:896–901.
23. Josephson G, Charbonneau G. Case report of reproductive problem in a new startup operation. J Swine Health Prod. 2001;9:258–259.
24. Johnson CS, Joo HS, Direksin K, Yoon KJ, Choi YK. Experimental in utero inoculation of late-term swine fetuses with porcine circovirus type 2. J Vet Diagn Invest. 2002;14:507–512.
25. O’Connor B, Gauvreau H, West K, Bogdan J, Ayroud M, Clark EG, Konoby C, Allan G, Ellis JA. Multiple porcine circovirus 2-associated abortions and reproductive failure in a multisite swine production unit. Can Vet J. 2001;42:551–553.
26. Ladekjær-Mikkelsen AS, Nielsen J, Storgaard T, Bøtner A, Allan G, McNeilly F. Transplacental infection with PCV-2 associated with reproductive failure in a gilt. Vet Rec. 2001;148:759–760.
27. Madson DM, Patterson AR, Ramamoorthy S, Pal N, Meng XJ, Opriessnig T. Effect of natural or vaccine-induced porcine circovirus type 2 (PCV2) immunity on fetal infection after artificial insemination with PCV2 spiked semen. Theriogenology. 2009;72:747–754.
*28. Cariolet R, Blanchard P, Dimna Ml, Truong C, Keranflec’h A, Beaurepaire B, Jolly JP, Julou P, Boisseson CD, Mahe D, Madec F, Jestin A. A study of different routes of inoculation of porcine circovirus type 2 (PCV2) to specific pathogen free (SPF) sows. Proc 34èmes Journées de la Recherche Porcin, Association Française de Zootechnie. Paris, France. 2002;317–323.
29. Hermann JR, Brockmeier SL, Yoon KJ, Zimmerman JJ. Detection of respiratory pathogens in air samples from acutely infected pigs. Can J Vet Res. 2008;72:367–370.
30. Cságola A, Cadar D, Tuboly T. Replication and transmission of porcine circovirus type 2 in mice. Acta Vet Hung. 2008;56:421–427.
31. Lorincz M, Cságola A, Biksi I, Szeredi L, Dán A, Tuboly T. Detection of porcine circovirus in rodents – Short communication. Acta Vet Hung. 2010;58:265–268.
32. Todd D. Avian circovirus diseases: lessons for the study of PMWS. Vet Microbiol. 2004;98:169–174.
33. Segalés J, Calsamiglia M, Olvera A, Sibila M, Badiella L, Domingo M. Quantification of porcine circovirus type 2 (PCV2) DNA in serum and tonsillar, nasal, tracheo-bronchial, urinary and faecal swabs of pigs with and without postweaning multisystemic wasting syndrome (PMWS). Vet Microbiol. 2005;111:223–229.
34. Opriessnig T, Prickett JR, Madson DM, Shen HG, Juhan NM, Pogranichniy R, Meng XJ, Halbur PG. Porcine circovirus type 2 (PCV2)-infection and re-inoculation with homologous or heterologous strains: Virological, serological, pathological and clinical effects in growing pigs. Vet Res. 2010;41:31. doi:10.1051/vetres/2010003.
35. Andraud M, Grasland B, Durand B, Cariolet R, Jestin A, Madec F, Rose N. Quantification of porcine circovirus type 2 (PCV-2) within- and between-pen transmission in pigs. Vet Res. 2008;39:43. doi:10.1051/vetres:2008020.
36. Madson DM, Patterson AR, Ramamoorthy S, Pal N, Meng XJ, Opriessnig T. Reproductive failure experimentally induced in sows via artificial insemination with semen spiked with porcine circovirus type 2. Vet Pathol. 2009;46:707–716.
*37. Sanford SE. Interesting diagnostic cases. Proc North Am Vet Conf Large Animal. Orlando, Florida. 2005;339–342.
38. Pejsak Z, Truszczynski M, Porowski M, PodgÓrska K. Effect of porcine circovirus type 2 vaccination of sows on selected production parameters. Bull Vet Inst Pulawy. 2009;53:159–164.
* Non-refereed references.
